R visualization: Distribution of longitudinal data
swimming plot
Introduction
Swimming plot shows the multiple characteristics of patients.
Loading required packages
knitr::opts_chunk$set(message = FALSE, warning = FALSE)
library(tidyverse)
library(ggtext)
# rm(list = ls())
options(stringsAsFactors = F)
Data preparation
clinic_data <- read.csv(paste0(getwd(), "/patients.csv"), row.names = 1)
head(clinic_data)
## PatientID Group Treatment Direction Type Day SampleID
## S_1 P01 Positive Treatment1 Right Yes 1 S_1
## S_2 P01 Positive Treatment1 Right Yes 67 S_2
## S_3 P01 Positive Treatment1 Right Yes 41 S_3
## S_4 P01 Positive Treatment1 Right Yes 85 S_4
## S_5 P01 Positive Treatment1 Right Yes 225 S_5
## S_6 P02 Positive Treatment2 Left No 0 S_6
Patients with clinical information have multiple samples during the therapeutic period. Here, we using swimming plot to show the distribution of sample per subject.
Plotting
Time Zone for intervals by Days
Factorizing the variables
plotdata <- clinic_data %>%
dplyr::select(PatientID, Group, Treatment, Direction, Type, Day, SampleID) %>%
dplyr::mutate(Day = as.numeric(Day)) %>%
dplyr::mutate(TimeZone = case_when(
Day > -30 & Day <= 21 ~ "T0",
Day > 21 & Day <= 90 ~ "T1",
Day > 91 & Day <= 180 ~ "T2",
Day > 181 & Day <= 270 ~ "T3",
Day > 271 ~ "T4")) %>%
dplyr::mutate(Group = factor(Group)) %>%
dplyr::mutate(Treatment = factor(Treatment)) %>%
dplyr::mutate(Direction = factor(Direction)) %>%
dplyr::mutate(Type = factor(Type)) %>%
dplyr::mutate(TimeZone = factor(TimeZone))
plotdata$PatientID <- factor(plotdata$PatientID, levels = rev(unique(plotdata$PatientID)))
# x scale
day_range <- as.integer(range(plotdata$Day))
MinDay <- day_range[1] - 2
MaxDay <- day_range[2] + 2
breaks_seq <- c(-30,
as.integer(rev(c(seq(0, MinDay, by=-10), MinDay))),
21,
as.integer(seq(60, MaxDay, by = 30)), MaxDay)
# y axis label
y_axis <- plotdata %>%
dplyr::select(PatientID, Group) %>%
dplyr::mutate(GroupCol = case_when(
Group == "Positive" ~ "#F28880",
Group == "Negative" ~ "#60C4D3"
)) %>%
dplyr::distinct() %>%
dplyr::mutate(PatientID = factor(PatientID, levels = levels(plotdata$PatientID)))
# Time Zone
Time_intervals <- data.frame(
xmin = c(-30, 21, 91, 181, 271),
xmax = c(21, 90, 180, 270, 360),
group = c("T0", "T1", "T2", "T3", "T4"))
- plotting
pl <- ggplot(plotdata) +
geom_point(aes(x = as.integer(Day), y = PatientID,
shape = Direction, color = Type), size = 3) +
geom_line(aes(x = as.integer(Day), y = PatientID,
color = Treatment)) +
geom_vline(xintercept = c(-30, 21, 90, 180, 270, 360),
color = "black", linewidth = 0.6, linetype = "dashed") +
scale_shape_manual(values = c(15, 16, 17)) +
scale_color_manual(values = c("red", "green", "blue",
"#32CD32", "#9400D3", "#48D1CC",
"#D51F26", "#272E6A", "#208A42", "#89288F")) +
geom_rect(data = Time_intervals, aes(xmin = xmin, xmax = xmax,
ymin = -Inf, ymax = Inf, fill = group),
alpha = 0.2) +
scale_fill_brewer(palette = "Spectral") +
labs(x = "Days", y = "PatientID") +
scale_x_continuous(breaks = breaks_seq) +
guides(
color = guide_legend(title = "Type & Treatment", order = 1),
shape = guide_legend(order = 2),
fill = guide_legend(title = "Time Zone", order = 3)) +
theme_bw() +
theme(axis.title = element_text(face = "bold", color = "black", size = 12),
axis.text.x = element_text(color = "black", size = 10),
axis.text.y = ggtext::element_markdown(size = 11, color = y_axis$GroupCol),
legend.key.size = unit(1.2, "line"),
legend.text = element_text(color = "black", size = 9))
pl
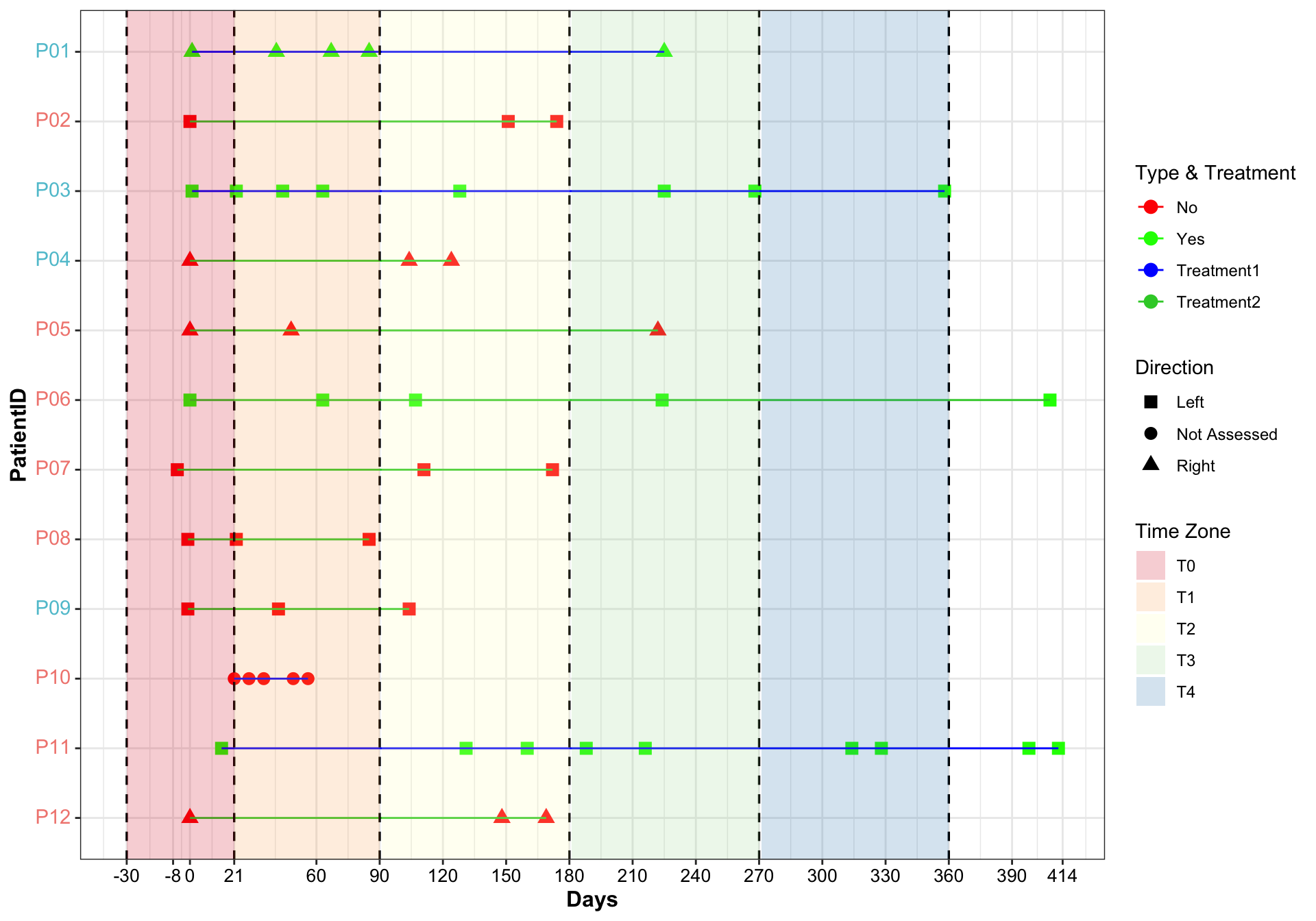
Results:
- y axis color: Group
Conclusion
Swimming plot presents lots of information from patients.
Systemic information
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.3.1 (2023-06-16)
## os macOS Monterey 12.2.1
## system x86_64, darwin20
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2024-03-14
## pandoc 3.1.3 @ /Users/zouhua/opt/anaconda3/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## blogdown 1.19 2024-02-01 [1] CRAN (R 4.3.2)
## bookdown 0.37 2023-12-01 [1] CRAN (R 4.3.0)
## bslib 0.6.1 2023-11-28 [1] CRAN (R 4.3.0)
## cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
## cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.0)
## colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
## commonmark 1.9.1 2024-01-30 [1] CRAN (R 4.3.2)
## devtools 2.4.5 2022-10-11 [1] CRAN (R 4.3.0)
## digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.0)
## dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.0)
## ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
## evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.0)
## fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.0)
## farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
## forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
## fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
## generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
## ggplot2 * 3.4.4 2023-10-12 [1] CRAN (R 4.3.0)
## ggtext * 0.1.2 2022-09-16 [1] CRAN (R 4.3.0)
## glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.0)
## gridtext 0.1.5 2022-09-16 [1] CRAN (R 4.3.0)
## gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
## highr 0.10 2022-12-22 [1] CRAN (R 4.3.0)
## hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
## htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.0)
## htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.0)
## httpuv 1.6.14 2024-01-26 [1] CRAN (R 4.3.2)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
## jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.0)
## knitr 1.45 2023-10-30 [1] CRAN (R 4.3.0)
## later 1.3.2 2023-12-06 [1] CRAN (R 4.3.0)
## lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.0)
## lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.0)
## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
## markdown 1.12 2023-12-06 [1] CRAN (R 4.3.0)
## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
## mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
## miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
## pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
## pkgbuild 1.4.3 2023-12-10 [1] CRAN (R 4.3.0)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
## pkgload 1.3.4 2024-01-16 [1] CRAN (R 4.3.0)
## profvis 0.3.8 2023-05-02 [1] CRAN (R 4.3.0)
## promises 1.2.1 2023-08-10 [1] CRAN (R 4.3.0)
## purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
## RColorBrewer 1.1-3 2022-04-03 [1] CRAN (R 4.3.0)
## Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.0)
## readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.0)
## remotes 2.4.2.1 2023-07-18 [1] CRAN (R 4.3.0)
## rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.0)
## rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.0)
## rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
## sass 0.4.8 2023-12-06 [1] CRAN (R 4.3.0)
## scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.0)
## sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
## shiny 1.8.0 2023-11-17 [1] CRAN (R 4.3.0)
## stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.0)
## stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.0)
## tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
## tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.2)
## tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
## tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
## timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.0)
## tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
## urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.3.0)
## usethis 2.2.2 2023-07-06 [1] CRAN (R 4.3.0)
## utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.0)
## vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.0)
## withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.0)
## xfun 0.41 2023-11-01 [1] CRAN (R 4.3.0)
## xml2 1.3.6 2023-12-04 [1] CRAN (R 4.3.0)
## xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
## yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────