R visualization: Raincloud plot by ggplot2
Raincloud plot
Introduction
Raincloud plot can be used to visualize raw data, the distribution of the data, and the key summary statistics at the same time.
Loading required packages
knitr::opts_chunk$set(message = FALSE, warning = FALSE)
library(ggpubr)
library(ggdist)
library(gghalves)
library(tidyverse)
# rm(list = ls())
options(stringsAsFactors = F)
# group & color
sp_names <- c("setosa", "versicolor", "virginica")
sp_colors <- c("#0073C2FF", "#EFC000FF", "#CD534CFF")
Data preparation
- Data object iris
data("iris")
data_pre <- iris
head(data_pre)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
Plot function
get_raincloud <- function(
dat,
group,
group_names,
group_colors,
measures) {
# group for plot x-label
dat_cln2 <- dat
colnames(dat_cln2)[which(colnames(dat_cln2) == group)] <- "Group_new"
if (group_names[1] == "all") {
tempdata <- dat_cln2
} else {
tempdata <- dat_cln2 %>%
dplyr::filter(Group_new %in% group_names)
}
tempdata$Group_new <- factor(tempdata$Group_new, levels = group_names)
if (length(measures) > 1) {
plotdata <- tempdata %>%
dplyr::select(all_of(c("Group_new", measures))) %>%
tidyr::pivot_longer(cols = 2:(length(measures)+1),
names_to = "Index",
values_to = "Values") %>%
dplyr::mutate(Index = factor(Index, levels = measures))
} else {
plotdata <- tempdata %>%
dplyr::select(all_of(c("Group_new", measures)))
colnames(plotdata)[2] <- "Values"
}
cmp <- list()
num <- utils::combn(length(group_names), 2)
for (i in 1:ncol(num)) {
cmp[[i]] <- num[, i]
}
if (length(measures) > 1) {
pl <- ggplot(plotdata, aes(x = Group_new, y = Values, fill = Group_new)) +
ggdist::stat_halfeye(adjust = 0.5, width = 0.3,
.width = 0, justification = -0.3, point_colour = NA) +
stat_boxplot(aes(color = Group_new), geom = "errorbar", width = 0.1) +
geom_boxplot(width = 0.1, outlier.shape = NA) +
gghalves::geom_half_point(side = "l", range_scale = 0.4, alpha = 0.5) +
stat_summary(geom = "crossbar", width = 0.08, fatten = 0, color = "white",
fun.data = function(x){c(y = median(x), ymin = median(x), ymax = median(x))}) +
labs(x = "", y = "Value") +
scale_fill_manual(values = group_colors) +
scale_color_manual(values = group_colors) +
guides(fill = "none", color = "none") +
scale_y_continuous(expand = expansion(mult = c(0.1, 0.1))) +
ggpubr::stat_compare_means(method = "wilcox.test",
comparisons = cmp) +
facet_wrap(.~ Index, scales = "free") +
theme_bw() +
theme(axis.title.y = element_text(size = 10, face = "bold"),
axis.text.y = element_text(size = 9),
axis.text.x = element_text(size = 10, hjust = .5, vjust = .5, angle = 30),
strip.text = element_text(size = 12, face = "bold", color = "black"))
} else {
pl <- ggplot(plotdata, aes(x = Group_new, y = Values, fill = Group_new)) +
ggdist::stat_halfeye(adjust = 0.5, width = 0.3,
.width = 0, justification = -0.3, point_colour = NA) +
stat_boxplot(aes(color = Group_new), geom = "errorbar", width = 0.1) +
geom_boxplot(width = 0.1, outlier.shape = NA) +
gghalves::geom_half_point(side = "l", range_scale = 0.4, alpha = 0.5) +
stat_summary(geom = "crossbar", width = 0.08, fatten = 0, color = "white",
fun.data = function(x){c(y = median(x), ymin = median(x), ymax = median(x))}) +
labs(x = "", y = measures) +
scale_fill_manual(values = group_colors) +
scale_color_manual(values = group_colors) +
guides(fill = "none", color = "none") +
scale_y_continuous(expand = expansion(mult = c(0.1, 0.1))) +
ggpubr::stat_compare_means(method = "wilcox.test",
comparisons = cmp) +
theme_bw() +
theme(axis.title.y = element_text(size = 10, face = "bold"),
axis.text.y = element_text(size = 9),
axis.text.x = element_text(size = 10, hjust = .5, vjust = .5, angle = 30),
strip.text = element_text(size = 12, face = "bold", color = "black"))
}
return(pl)
}
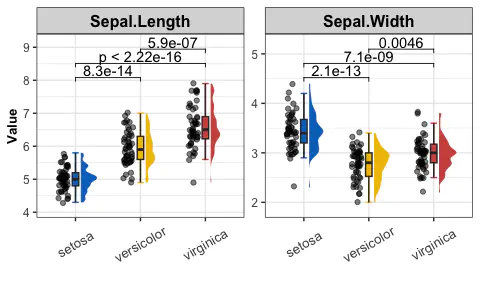
Raincloud plot with single measure
get_raincloud(
dat = data_pre,
group = "Species",
group_names = sp_names,
group_colors = sp_colors,
measures = "Sepal.Length")
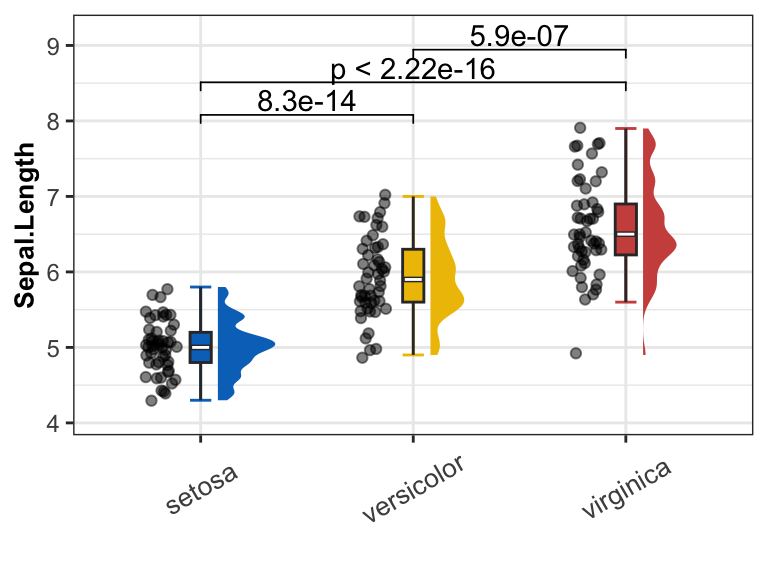
Results:
- Raincloud plot shows the distribution of the data and the boxplot of data in three groups.
Raincloud plot with mulitple measures
get_raincloud(
dat = data_pre,
group = "Species",
group_names = sp_names,
group_colors = sp_colors,
measures = c("Sepal.Length", "Sepal.Width"))
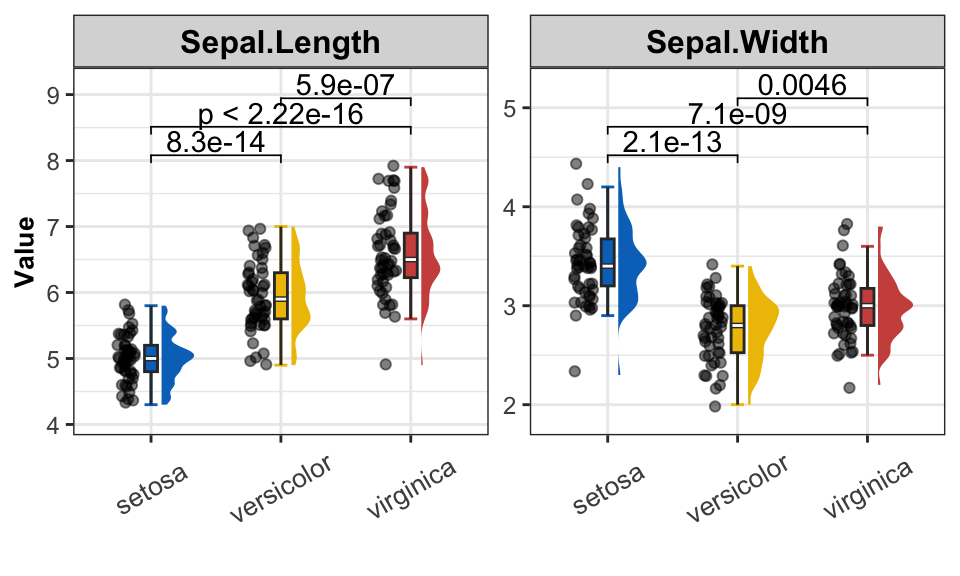
Conclusion
RainCloud plots using Half Violin plot with jittered data points to show the distribution of the data.
Systemic information
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.3.1 (2023-06-16)
## os macOS Monterey 12.2.1
## system x86_64, darwin20
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2024-03-14
## pandoc 3.1.3 @ /Users/zouhua/opt/anaconda3/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## abind 1.4-5 2016-07-21 [1] CRAN (R 4.3.0)
## backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
## blogdown 1.19 2024-02-01 [1] CRAN (R 4.3.2)
## bookdown 0.37 2023-12-01 [1] CRAN (R 4.3.0)
## broom 1.0.5 2023-06-09 [1] CRAN (R 4.3.0)
## bslib 0.6.1 2023-11-28 [1] CRAN (R 4.3.0)
## cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
## car 3.1-2 2023-03-30 [1] CRAN (R 4.3.0)
## carData 3.0-5 2022-01-06 [1] CRAN (R 4.3.0)
## cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.0)
## colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
## devtools 2.4.5 2022-10-11 [1] CRAN (R 4.3.0)
## digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.0)
## distributional 0.3.2 2023-03-22 [1] CRAN (R 4.3.0)
## dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.0)
## ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
## evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.0)
## fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.0)
## farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
## forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
## fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
## generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
## ggdist * 3.3.1 2023-11-27 [1] CRAN (R 4.3.0)
## gghalves * 0.1.4 2022-11-20 [1] CRAN (R 4.3.0)
## ggplot2 * 3.4.4 2023-10-12 [1] CRAN (R 4.3.0)
## ggpubr * 0.6.0 2023-02-10 [1] CRAN (R 4.3.0)
## ggsignif 0.6.4 2022-10-13 [1] CRAN (R 4.3.0)
## glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.0)
## gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
## highr 0.10 2022-12-22 [1] CRAN (R 4.3.0)
## hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
## htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.0)
## htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.0)
## httpuv 1.6.14 2024-01-26 [1] CRAN (R 4.3.2)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
## jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.0)
## knitr 1.45 2023-10-30 [1] CRAN (R 4.3.0)
## labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
## later 1.3.2 2023-12-06 [1] CRAN (R 4.3.0)
## lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.0)
## lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.0)
## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
## mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
## miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
## pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
## pkgbuild 1.4.3 2023-12-10 [1] CRAN (R 4.3.0)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
## pkgload 1.3.4 2024-01-16 [1] CRAN (R 4.3.0)
## profvis 0.3.8 2023-05-02 [1] CRAN (R 4.3.0)
## promises 1.2.1 2023-08-10 [1] CRAN (R 4.3.0)
## purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
## Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.0)
## readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.0)
## remotes 2.4.2.1 2023-07-18 [1] CRAN (R 4.3.0)
## rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.0)
## rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.0)
## rstatix 0.7.2 2023-02-01 [1] CRAN (R 4.3.0)
## rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
## sass 0.4.8 2023-12-06 [1] CRAN (R 4.3.0)
## scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.0)
## sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
## shiny 1.8.0 2023-11-17 [1] CRAN (R 4.3.0)
## stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.0)
## stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.0)
## tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
## tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.2)
## tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
## tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
## timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.0)
## tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
## urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.3.0)
## usethis 2.2.2 2023-07-06 [1] CRAN (R 4.3.0)
## utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.0)
## vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.0)
## withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.0)
## xfun 0.41 2023-11-01 [1] CRAN (R 4.3.0)
## xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
## yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────