R visualization: barplot with chisqure test by ggplot2
grouped barplot with chisqure test
Introduction
chi-square test is used to test whether there is a significant difference between categories. Here, we combined barplot and chi-square test to display these results.
Loading required packages
knitr::opts_chunk$set(message = FALSE, warning = FALSE)
library(tidyverse)
# install.packages("titanic")
library(titanic)
# rm(list = ls())
options(stringsAsFactors = F)
Data preparation
- Data object from titanic R package
dat <- as_tibble(titanic_train)
head(dat)
## # A tibble: 6 × 12
## PassengerId Survived Pclass Name Sex Age SibSp Parch Ticket Fare Cabin
## <int> <int> <int> <chr> <chr> <dbl> <int> <int> <chr> <dbl> <chr>
## 1 1 0 3 Braund… male 22 1 0 A/5 2… 7.25 ""
## 2 2 1 1 Cuming… fema… 38 1 0 PC 17… 71.3 "C85"
## 3 3 1 3 Heikki… fema… 26 0 0 STON/… 7.92 ""
## 4 4 1 1 Futrel… fema… 35 1 0 113803 53.1 "C12…
## 5 5 0 3 Allen,… male 35 0 0 373450 8.05 ""
## 6 6 0 3 Moran,… male NA 0 0 330877 8.46 ""
## # ℹ 1 more variable: Embarked <chr>
- Factorizing Group
plotdata <- dat %>%
dplyr::filter(Pclass == 1) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::group_by(Sex, Survived) %>%
dplyr::summarise(value = n()) %>%
dplyr::mutate(Pclass = "Pclass1") %>%
dplyr::ungroup() %>%
rbind(dat %>%
dplyr::filter(Pclass == 2) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::group_by(Sex, Survived) %>%
dplyr::summarise(value = n()) %>%
dplyr::mutate(Pclass = "Pclass2") %>%
dplyr::ungroup()
) %>%
rbind(dat %>%
dplyr::filter(Pclass == 3) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::group_by(Sex, Survived) %>%
dplyr::summarise(value = n()) %>%
dplyr::mutate(Pclass = "Pclass3") %>%
dplyr::ungroup()
) %>%
dplyr::mutate(Sex = factor(Sex, levels = c("male", "female"))) %>%
dplyr::mutate(Survived = factor(Survived, levels = c("0", "1"), labels = c("No", "Yes"))) %>%
dplyr::mutate(group = paste(Sex, Survived)) %>%
dplyr::mutate(Pclass = factor(Pclass, levels = c("Pclass1", "Pclass2", "Pclass3"))) %>%
dplyr::mutate(group = factor(group, levels = c("male No", "female No", "male Yes", "female Yes")))
head(plotdata)
## # A tibble: 6 × 5
## Sex Survived value Pclass group
## <fct> <fct> <int> <fct> <fct>
## 1 female No 3 Pclass1 female No
## 2 female Yes 91 Pclass1 female Yes
## 3 male No 77 Pclass1 male No
## 4 male Yes 45 Pclass1 male Yes
## 5 female No 6 Pclass2 female No
## 6 female Yes 70 Pclass2 female Yes
- test by
chisq.test
test_res <- dat %>%
dplyr::filter(Pclass == 1) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::mutate(Pclass = "Pclass1") %>%
rbind(dat %>%
dplyr::filter(Pclass == 2) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::mutate(Pclass = "Pclass2")
) %>%
rbind(dat %>%
dplyr::filter(Pclass == 3) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::mutate(Pclass = "Pclass3")
) %>%
dplyr::group_by(Pclass) %>%
dplyr::summarise(statistic = chisq.test(table(Sex, Survived))$statistic,
parameter = chisq.test(table(Sex, Survived))$parameter,
p.value = signif(chisq.test(table(Sex, Survived))$p.value, 3)) %>%
dplyr::ungroup() %>%
dplyr::mutate(Pclass = factor(Pclass, levels = c("Pclass1", "Pclass2", "Pclass3")))
test_res
## # A tibble: 3 × 4
## Pclass statistic parameter p.value
## <fct> <dbl> <int> <dbl>
## 1 Pclass1 79.2 1 5.6 e-19
## 2 Pclass2 101. 1 7.82e-24
## 3 Pclass3 71.7 1 2.53e-17
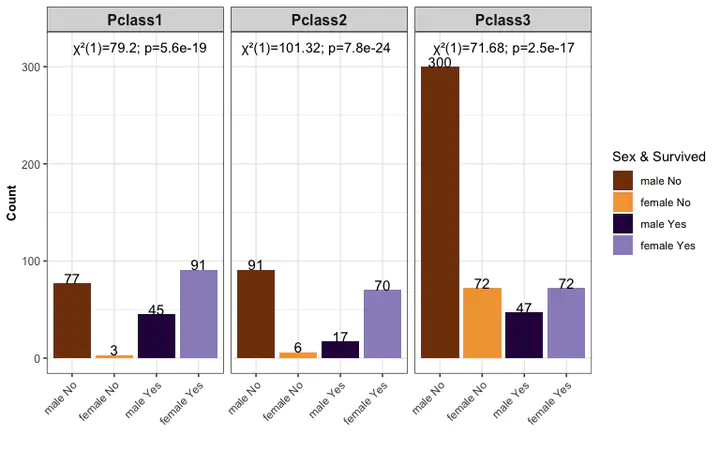
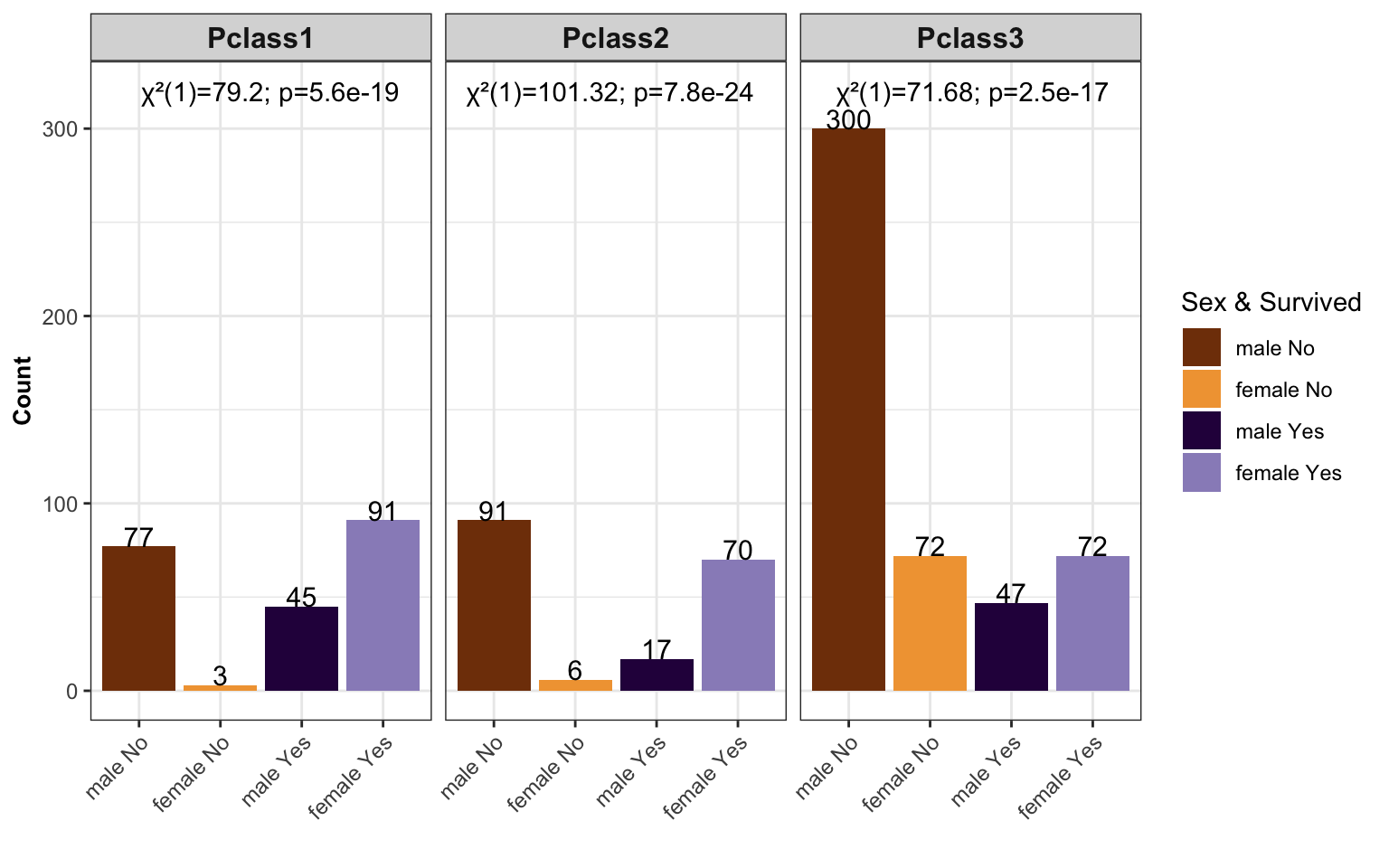
Bar Plot with Chi-square test
- The chi-square test is the kind of hypothesis test that we use to examine the relationship between two categorical variables.
barpl <- ggplot(data = plotdata) +
geom_bar(aes(x = group, y = value, fill = group), stat = "identity", position = position_dodge()) +
geom_text(aes(x = group, y = value, label = value), vjust = 0, color = "black", size = 4) +
geom_text(data = test_res,
aes(x = 4.2, y = max(plotdata$value) + 20,
label = paste0("χ²(", parameter, ")=", round(statistic, 2), "; p=", signif(p.value, 2))),
hjust = 1) +
labs(x = "", y = "Count") +
scale_fill_manual(name = "Sex & Survived",
values = c("#803C08", "#F1A340",
"#2C0a4B", "#998EC3")) +
facet_grid(.~ Pclass, scales = "free_x") +
theme_bw() +
theme(axis.title.y = element_text(size = 10, face = "bold"),
axis.text.y = element_text(size = 9),
axis.text.x = element_text(angle = 45, hjust = 1, size = 9),
strip.text = element_text(size = 12, face = "bold"))
barpl
- stacked barplot
plotdata2 <- dat %>%
dplyr::filter(Pclass == 1) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::group_by(Sex, Survived) %>%
dplyr::summarise(value = n()) %>%
dplyr::mutate(Pclass = "Pclass1") %>%
dplyr::ungroup() %>%
rbind(dat %>%
dplyr::filter(Pclass == 2) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::group_by(Sex, Survived) %>%
dplyr::summarise(value = n()) %>%
dplyr::mutate(Pclass = "Pclass2") %>%
dplyr::ungroup()
) %>%
rbind(dat %>%
dplyr::filter(Pclass == 3) %>%
dplyr::select(PassengerId, Sex, Survived) %>%
dplyr::group_by(Sex, Survived) %>%
dplyr::summarise(value = n()) %>%
dplyr::mutate(Pclass = "Pclass3") %>%
dplyr::ungroup()
) %>%
dplyr::mutate(Sex = factor(Sex, levels = c("male", "female"))) %>%
dplyr::mutate(Survived = factor(Survived, levels = c("0", "1"), labels = c("No", "Yes"))) %>%
dplyr::arrange(desc(Survived)) %>%
dplyr::group_by(Pclass, Sex) %>%
dplyr::mutate(percent = signif(value/sum(value), 4),
pos = cumsum(percent) - 0.5*percent,
label = paste0(percent * 100, "% (", value, ")"))
stacked_barpl <- ggplot(data = plotdata2, aes(x = Sex, y = percent)) +
geom_bar(aes(fill = Survived),
stat = "identity", position = "fill", color = "black") +
geom_text(aes(label = ifelse(percent >= 0.07, label,""),
y = pos), color = "white", size = 3.5) +
geom_text(data = test_res,
aes(x = 2.2, y = 1.1,
label = paste0("χ²(", parameter, ")=", round(statistic, 2), "; p=", signif(p.value, 2))),
hjust = 1) +
labs(x = "", y = "Count (%)") +
scale_fill_manual(name = "Survived",
values = c("#803C08", "#F1A340")) +
# scale_y_continuous(breaks = seq(0, 1, 0.2),
# labels = paste0(seq(0, 1, 0.2) * 100, "%"),
# expand = expansion(mult = c(0, 0.1))) +
facet_grid(.~ Pclass, scales = "free_x") +
theme_bw() +
theme(axis.title.y = element_text(size = 10, face = "bold"),
axis.text.y = element_text(size = 9),
axis.text.x = element_text(angle = 45, hjust = 1, size = 9),
strip.text = element_text(size = 12, face = "bold"))
stacked_barpl
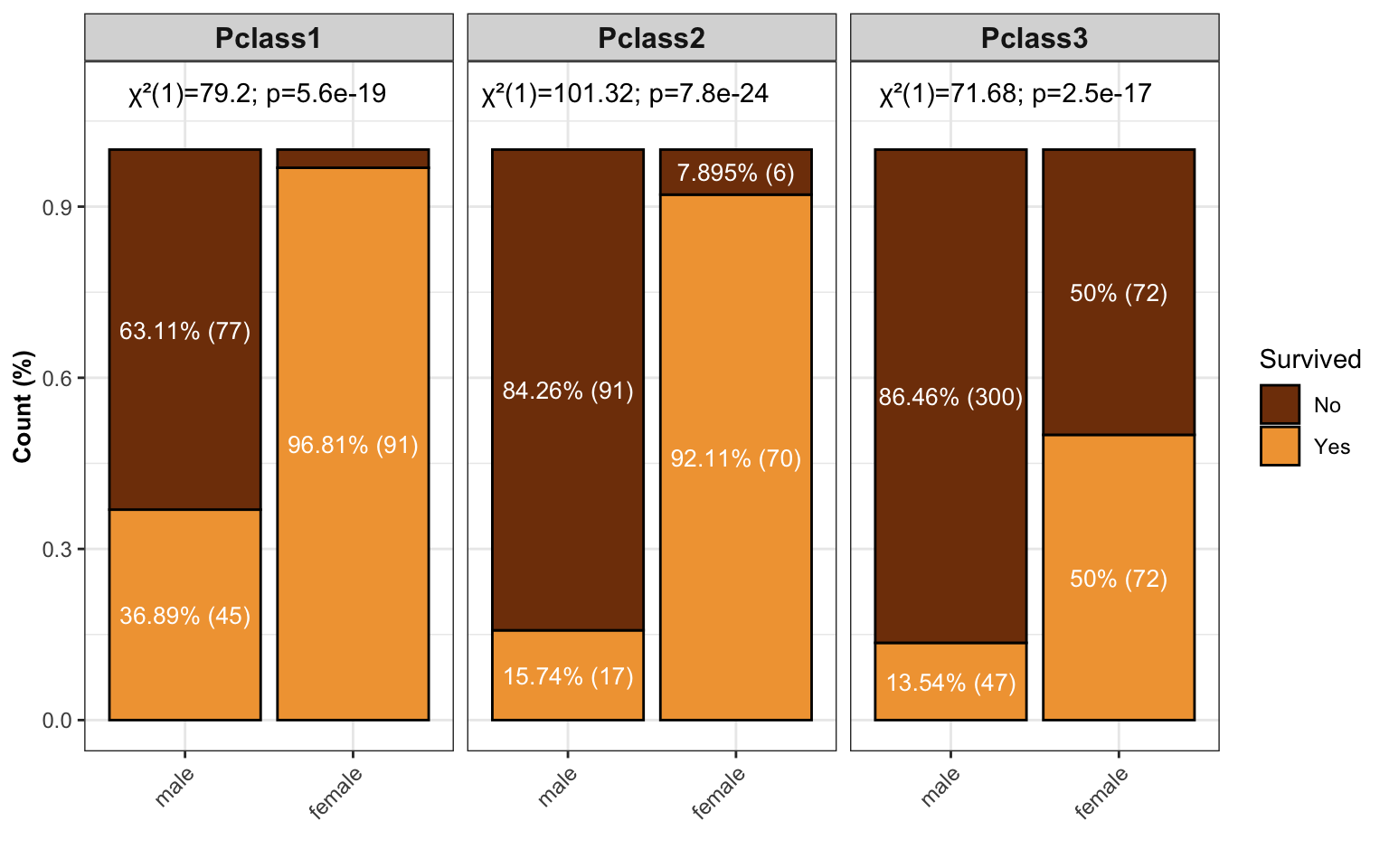
Conclusion
barplot shows the number of groups and add chi-square test into the barplot.
Systemic information
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.3 (2022-03-10)
## os macOS Big Sur/Monterey 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2024-01-22
## pandoc 3.1.3 @ /Users/zouhua/opt/anaconda3/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## blogdown 1.18 2023-06-19 [2] CRAN (R 4.1.3)
## bookdown 0.34 2023-05-09 [2] CRAN (R 4.1.2)
## bslib 0.6.0 2023-11-21 [1] CRAN (R 4.1.3)
## cachem 1.0.8 2023-05-01 [2] CRAN (R 4.1.2)
## callr 3.7.3 2022-11-02 [2] CRAN (R 4.1.2)
## cli 3.6.1 2023-03-23 [2] CRAN (R 4.1.2)
## colorspace 2.1-0 2023-01-23 [2] CRAN (R 4.1.2)
## crayon 1.5.2 2022-09-29 [2] CRAN (R 4.1.2)
## devtools 2.4.5 2022-10-11 [2] CRAN (R 4.1.2)
## digest 0.6.33 2023-07-07 [1] CRAN (R 4.1.3)
## dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.1.3)
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.1.0)
## evaluate 0.21 2023-05-05 [2] CRAN (R 4.1.2)
## fansi 1.0.4 2023-01-22 [2] CRAN (R 4.1.2)
## farver 2.1.1 2022-07-06 [2] CRAN (R 4.1.2)
## fastmap 1.1.1 2023-02-24 [2] CRAN (R 4.1.2)
## forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.1.2)
## fs 1.6.2 2023-04-25 [2] CRAN (R 4.1.2)
## generics 0.1.3 2022-07-05 [2] CRAN (R 4.1.2)
## ggplot2 * 3.4.4 2023-10-12 [1] CRAN (R 4.1.3)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.1.2)
## gtable 0.3.3 2023-03-21 [2] CRAN (R 4.1.2)
## highr 0.10 2022-12-22 [2] CRAN (R 4.1.2)
## hms 1.1.3 2023-03-21 [2] CRAN (R 4.1.2)
## htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.1.3)
## htmlwidgets 1.6.2 2023-03-17 [2] CRAN (R 4.1.2)
## httpuv 1.6.11 2023-05-11 [2] CRAN (R 4.1.3)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.1.0)
## jsonlite 1.8.7 2023-06-29 [2] CRAN (R 4.1.3)
## knitr 1.43 2023-05-25 [2] CRAN (R 4.1.3)
## labeling 0.4.2 2020-10-20 [2] CRAN (R 4.1.0)
## later 1.3.1 2023-05-02 [2] CRAN (R 4.1.2)
## lifecycle 1.0.3 2022-10-07 [2] CRAN (R 4.1.2)
## lubridate * 1.9.2 2023-02-10 [2] CRAN (R 4.1.2)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.1.2)
## memoise 2.0.1 2021-11-26 [2] CRAN (R 4.1.0)
## mime 0.12 2021-09-28 [2] CRAN (R 4.1.0)
## miniUI 0.1.1.1 2018-05-18 [2] CRAN (R 4.1.0)
## munsell 0.5.0 2018-06-12 [2] CRAN (R 4.1.0)
## pillar 1.9.0 2023-03-22 [2] CRAN (R 4.1.2)
## pkgbuild 1.4.2 2023-06-26 [2] CRAN (R 4.1.3)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.1.0)
## pkgload 1.3.2.1 2023-07-08 [2] CRAN (R 4.1.3)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.1.0)
## processx 3.8.2 2023-06-30 [2] CRAN (R 4.1.3)
## profvis 0.3.8 2023-05-02 [2] CRAN (R 4.1.2)
## promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.1.0)
## ps 1.7.5 2023-04-18 [2] CRAN (R 4.1.2)
## purrr * 1.0.1 2023-01-10 [1] CRAN (R 4.1.2)
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.1.0)
## Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.1.3)
## readr * 2.1.4 2023-02-10 [1] CRAN (R 4.1.2)
## remotes 2.4.2 2021-11-30 [2] CRAN (R 4.1.0)
## rlang 1.1.1 2023-04-28 [1] CRAN (R 4.1.2)
## rmarkdown 2.23 2023-07-01 [2] CRAN (R 4.1.3)
## rstudioapi 0.15.0 2023-07-07 [2] CRAN (R 4.1.3)
## sass 0.4.6 2023-05-03 [2] CRAN (R 4.1.2)
## scales 1.2.1 2022-08-20 [1] CRAN (R 4.1.2)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.1.0)
## shiny 1.7.4.1 2023-07-06 [2] CRAN (R 4.1.3)
## stringi 1.7.12 2023-01-11 [2] CRAN (R 4.1.2)
## stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.1.3)
## tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.1.2)
## tidyr * 1.3.0 2023-01-24 [1] CRAN (R 4.1.2)
## tidyselect 1.2.0 2022-10-10 [2] CRAN (R 4.1.2)
## tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.1.2)
## timechange 0.2.0 2023-01-11 [2] CRAN (R 4.1.2)
## titanic * 0.1.0 2015-08-31 [1] CRAN (R 4.1.0)
## tzdb 0.4.0 2023-05-12 [2] CRAN (R 4.1.3)
## urlchecker 1.0.1 2021-11-30 [2] CRAN (R 4.1.0)
## usethis 2.2.2 2023-07-06 [2] CRAN (R 4.1.3)
## utf8 1.2.3 2023-01-31 [2] CRAN (R 4.1.2)
## vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.1.3)
## withr 2.5.0 2022-03-03 [2] CRAN (R 4.1.2)
## xfun 0.40 2023-08-09 [1] CRAN (R 4.1.3)
## xtable 1.8-4 2019-04-21 [2] CRAN (R 4.1.0)
## yaml 2.3.7 2023-01-23 [2] CRAN (R 4.1.2)
##
## [1] /Users/zouhua/Library/R/x86_64/4.1/library
## [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────