R visualization: scatter plot on PCA by ggplot2
scatter plot on PCA
Introduction
Display the results of PCA (Principle Components Analysis) by using a scatterplot. It’s also necessary to draw the boundaries of the group. Here, this tutorial would give four approaches to do this using ggplot2.
Loading required packages
knitr::opts_chunk$set(message = FALSE, warning = FALSE)
library(tidyverse)
library(patchwork)
# rm(list = ls())
options(stringsAsFactors = F)
# group & color
group_names <- c("A", "B", "C")
group_colors <- c("#0073C2FF", "#EFC000FF", "#CD534CFF")
group_shapes <- c(15, 16, 17)
Data preparation
- Constructing data object
dat <- data.frame(
SampleID = paste0("S_", 1:24),
PC1 = c(0.022770707, 0.054692482, 0.039981067, 0.007793265, 7.84e-06, -0.009869502, -0.059123147,
-0.100341862, 0.027247694, 0.156717826, 0.247041497, 0.181258984, 0.172786176,
0.131875865, 0.230052533, 0.163389878, -0.116222295, -0.221050628, -0.084606354,
-0.214705258, -0.210235971, -0.200839043, -0.06763774, -0.175523677),
PC2 = c(-0.05555032, -0.050592178, -0.070996307, -0.037754744, -0.091904909, -0.067078552,
-0.114239498, -0.099288661, 0.024239885, 0.06885023, 0.005302438, 0.035700696, 0.042451004,
0.019859585, 0.019250596, 0.031094568, 0.022300445, 0.057798633, 0.08757355, 0.048752276,
0.021344081, 0.055314429, -0.005297042, 0.10518067),
PC3 = c(-0.052194396, 0.000958851, 0.006141922, 0.048682841, -0.061645805, -0.003102679, 0.028396414,
0.01980974, -0.055276428, -0.082034627, 0.081183975, 0.030190352, -0.035916706, -0.052318759,
0.016482896, 0.096296503, -0.01270539, 0.019406331, 0.033471156, 0.003175988, 0.020986428,
0.025259074, -0.093440324, 0.062841498),
Group = c(rep("A", 8), rep("B", 8), rep("C", 8)),
PID = paste0("P", rep(1:8, 3)))
Notice:
SampleID: sample names
PC1: the first principle component from PCA
PC2: the second principle component from PCA
PC3: the third principle component from PCA
Group: group information
PID: participant identity
- Factorizing Group
plotdata <- dat |>
dplyr::mutate(Group = factor(Group, levels = group_names))
head(plotdata)
## SampleID PC1 PC2 PC3 Group PID
## 1 S_1 0.022770707 -0.05555032 -0.052194396 A P1
## 2 S_2 0.054692482 -0.05059218 0.000958851 A P2
## 3 S_3 0.039981067 -0.07099631 0.006141922 A P3
## 4 S_4 0.007793265 -0.03775474 0.048682841 A P4
## 5 S_5 0.000007840 -0.09190491 -0.061645805 A P5
## 6 S_6 -0.009869502 -0.06707855 -0.003102679 A P6
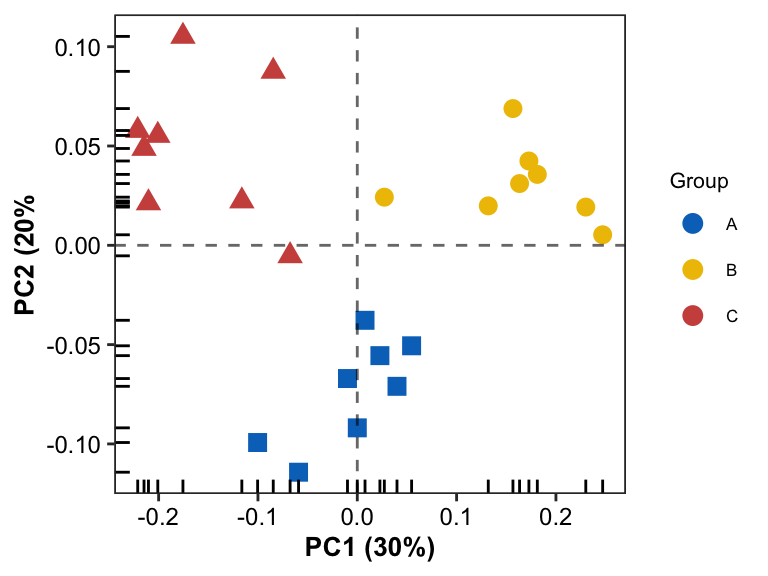
Simple Scatter Plot
- create poing by
geom_point
pl0 <- ggplot(data = plotdata, aes(x = PC1, y = PC2)) +
geom_point(aes(color = Group, shape = Group), size = 3) +
geom_rug() +
geom_hline(yintercept = 0, linetype = 2, linewidth = 0.5, alpha = 0.6) +
geom_vline(xintercept = 0, linetype = 2, linewidth = 0.5, alpha = 0.6) +
labs(x = "PC1 (30%)", y = "PC2 (20%") +
scale_color_manual(values = group_colors) +
scale_shape_manual(values = group_shapes) +
guides(shape = "none") +
theme_bw() +
theme(plot.title = element_text(size = 10, color = "black", face = "bold", hjust = 0.5),
axis.title = element_text(size = 10, color = "black", face = "bold"),
axis.text = element_text(size = 9, color = "black"),
text = element_text(size = 8, color = "black"),
strip.text = element_text(size = 9, color = "black", face = "bold"),
panel.grid = element_blank(),
panel.background = element_rect(color = "black", fill = "transparent"),
legend.key = element_rect(fill = "transparent"))
pl0
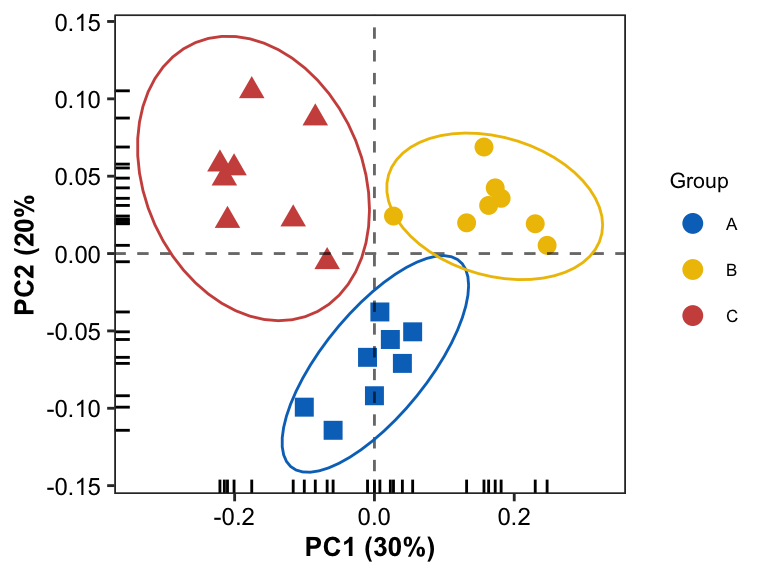
Scatter Plot with confidence interval of group
- add confidence interval ellipse by
stat_ellipse
pl1 <- pl0 + stat_ellipse(aes(color = Group), level = 0.95, linetype = 1, show.legend = FALSE)
pl1
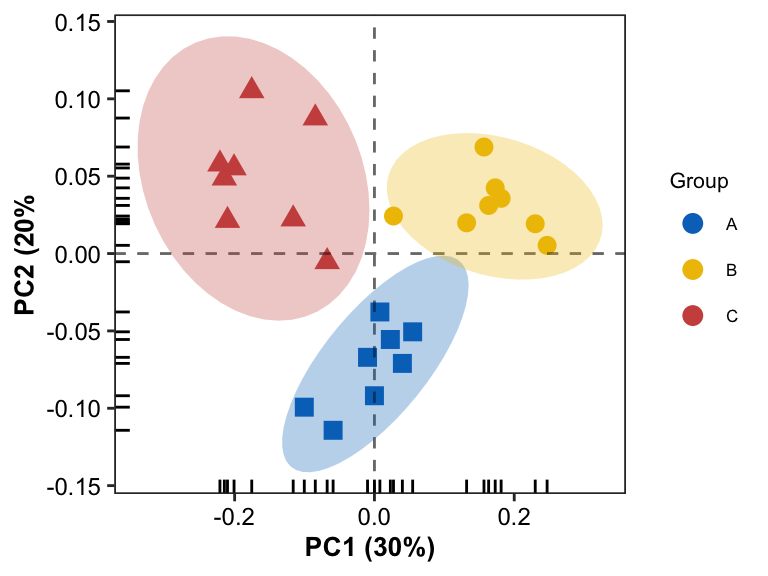
Another Scatter Plot with confidence interval
- add confidence interval ellipse by
stat_ellipse
pl2 <- pl0 + stat_ellipse(aes(fill = Group), geom = "polygon", level = 0.95, alpha = 0.3, show.legend = FALSE) +
scale_fill_manual(values = group_colors)
pl2
Scatter Plot with enterotype
- Add enterotype by
geom_enterotype
# https://github.com/cran/miscTools/
insertRow <- function(m, r, v = NA, rName = "") {
if (!inherits(m, "matrix")) {
stop("argument 'm' must be a matrix")
}
if (r == as.integer(r)) {
r <- as.integer(r)
}
else {
stop("argument 'r' must be an integer")
}
if (length(r) != 1) {
stop("argument 'r' must be a scalar")
}
if (r < 1) {
stop("argument 'r' must be positive")
}
if (r > nrow(m) + 1) {
stop("argument 'r' must not be larger than the number of rows",
" of matrix 'm' plus one")
}
if (!is.character(rName)) {
stop("argument 'rName' must be a character string")
}
if (length(rName) != 1) {
stop("argument 'rName' must be a be a single character string")
}
nr <- nrow(m)
nc <- ncol(m)
rNames <- rownames(m)
if (is.null(rNames) & rName != "") {
rNames <- rep("", nr)
}
if (r == 1) {
m2 <- rbind(matrix(v, ncol = nc), m)
if (!is.null(rNames)) {
rownames(m2) <- c(rName, rNames)
}
}
else if (r == nr + 1) {
m2 <- rbind(m, matrix(v, ncol = nc))
if (!is.null(rNames)) {
rownames(m2) <- c(rNames, rName)
}
}
else {
m2 <- rbind(m[1:(r - 1), , drop = FALSE], matrix(v, ncol = nc),
m[r:nr, , drop = FALSE])
if (!is.null(rNames)) {
rownames(m2) <- c(rNames[1:(r - 1)], rName, rNames[r:nr])
}
}
return(m2)
}
# https://stackoverflow.com/questions/42575769/how-to-modify-the-backgroup-color-of-label-in-the-multiple-ggproto-using-ggplot2
geom_enterotype <- function(
mapping = NULL,
data = NULL,
stat = "identity",
position = "identity",
alpha = 0.2,
prop = 0.5,
lineend = "butt",
linejoin = "round",
linemitre = 1,
arrow = NULL,
na.rm = FALSE,
parse = FALSE,
nudge_x = 0,
nudge_y = 0,
label.padding = unit(0.15, "lines"),
label.r = unit(0.15, "lines"),
label.size = 0.1,
show.legend = TRUE,
inherit.aes = TRUE,
...) {
# create new stat and geom for scatterplot with ellipses
StatEllipse <- ggproto(
"StatEllipse",
Stat,
required_aes = c("x", "y"),
compute_group = function(., data, scales, level = 0.75, segments = 51, ...) {
#library(MASS)
dfn <- 2
dfd <- length(data$x) - 1
if (dfd < 3) {
ellipse <- rbind(c(NA, NA))
} else {
v <- MASS::cov.trob(cbind(data$x, data$y))
shape <- v$cov
center <- v$center
radius <- sqrt(dfn * qf(level, dfn, dfd))
angles <- (0:segments) * 2 * pi / segments
unit.circle <- cbind(cos(angles), sin(angles))
ellipse <- t(center + radius * t(unit.circle %*% chol(shape)))
}
ellipse <- as.data.frame(ellipse)
colnames(ellipse) <- c("x", "y")
return(ellipse)
}
)
# write new ggproto
GeomEllipse <- ggproto(
"GeomEllipse",
Geom,
draw_group = function(data, panel_scales, coord) {
n <- nrow(data)
if (n == 1) {
return(zeroGrob())
}
munched <- coord_munch(coord, data, panel_scales)
munched <- munched[order(munched$group), ]
first_idx <- !duplicated(munched$group)
first_rows <- munched[first_idx, ]
grid::pathGrob(munched$x, munched$y, default.units = "native",
id = munched$group,
gp = grid::gpar(col = first_rows$colour,
fill = alpha(first_rows$fill, first_rows$alpha),
lwd = first_rows$size * .pt,
lty = first_rows$linetype))
}, default_aes = aes(colour = "NA", fill = "grey20", size = 0.5,
linetype = 1, alpha = NA, prop = 0.5),
handle_na = function(data, params) {data},
required_aes = c("x", "y"),
draw_key = draw_key_path
)
# create a new stat for PCA scatterplot with lines which totally directs to the center
StatConline <- ggproto(
"StatConline",
Stat,
compute_group = function(data, scales) {
df <- data.frame(data$x, data$y)
mat <- as.matrix(df)
center <- MASS::cov.trob(df)$center
names(center) <- NULL
mat_insert <- insertRow(mat, 2, center)
for(i in 1:nrow(mat)) {
mat_insert <- insertRow(mat_insert, 2*i, center)
next
}
mat_insert <- mat_insert[-c(2:3), ]
rownames(mat_insert) <- NULL
mat_insert <- as.data.frame(mat_insert, center)
colnames(mat_insert) <- c("x", "y")
return(mat_insert)
}, required_aes = c("x", "y")
)
# create a new stat for PCA scatterplot with center labels
StatLabel <- ggproto(
"StatLabel",
Stat,
compute_group = function(data, scales) {
df <- data.frame(data$x, data$y)
center <- MASS::cov.trob(df)$center
names(center) <- NULL
center <- t(as.data.frame(center))
center <- as.data.frame(cbind(center))
colnames(center) <- c("x", "y")
rownames(center) <- NULL
return(center)
}, required_aes = c("x", "y")
)
layer1 <- layer(data = data,
mapping = mapping,
stat = stat,
geom = GeomPoint,
position = position,
show.legend = show.legend,
inherit.aes = inherit.aes,
params = list(na.rm = na.rm, ...))
layer2 <- layer(data = data,
mapping = mapping,
stat = StatEllipse,
geom = GeomEllipse,
position = position,
show.legend = FALSE,
inherit.aes = inherit.aes,
params = list(na.rm = na.rm, prop = prop, alpha = alpha, ...))
layer3 <- layer(data = data,
mapping = mapping,
stat = StatConline,
geom = GeomPath,
position = position,
show.legend = show.legend,
inherit.aes = inherit.aes,
params = list(lineend = lineend, linejoin = linejoin,
linemitre = linemitre, arrow = arrow, na.rm = na.rm, ...))
if (!missing(nudge_x) || !missing(nudge_y)) {
if (!missing(position)) {
stop("Specify either `position` or `nudge_x`/`nudge_y`",
call. = FALSE)
}
position <- position_nudge(nudge_x, nudge_y)
}
layer4 <- layer(data = data,
mapping = mapping,
stat = StatLabel,
geom = GeomLabel,
position = position,
show.legend = FALSE,
inherit.aes = inherit.aes,
params = list(parse = parse, label.padding = label.padding,
label.r = label.r, label.size = label.size, na.rm = na.rm, ...))
return(list(layer1, layer2, layer3, layer4))
}
- plotting
pl3 <- pl0 + geom_enterotype(aes(color = Group, label = Group), show.legend = FALSE, alpha = 0.1)
pl3
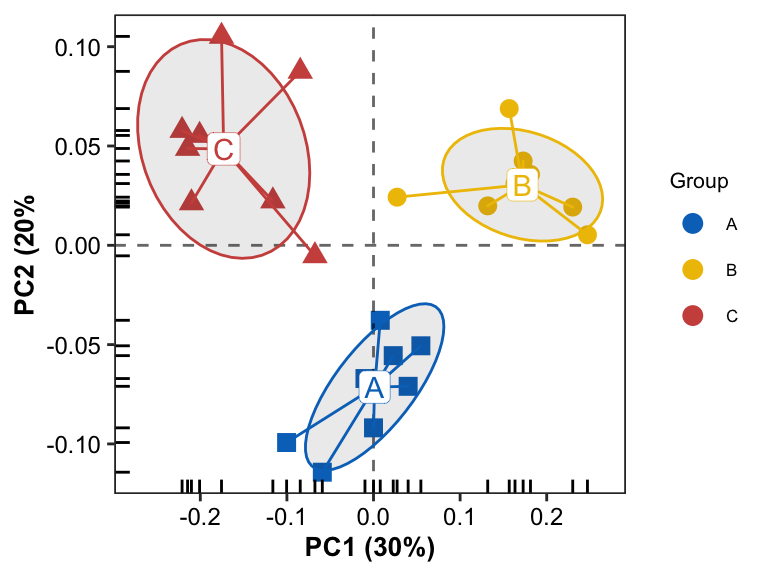
Scatter Plot with the border line
Calculating the position of group label
Calculating the position of outlier per group
Compvar_position <- cbind(PC1 = tapply(plotdata$PC1, plotdata$Group, mean),
PC2 = tapply(plotdata$PC2, plotdata$Group, mean)) %>%
as.data.frame() %>%
tibble::rownames_to_column("Group")
border_fun <- function(x = plotdata, grp) {
tempdat <- x[x$Group %in% grp, , F]
res <- tempdat[chull(tempdat[[2]], tempdat[[3]]), ]
return(res)
}
Compvar_border <- NULL
for (i in 1:length(unique(plotdata$Group))) {
Compvar_border <- rbind(Compvar_border, border_fun(grp = unique(plotdata$Group)[i]))
}
- Plotting
pl4 <- pl0 + geom_line(aes(group = PID), linetype = 2, alpha = 0.2) +
geom_text(data = Compvar_position, aes(x = PC1, y = PC2, label = Group), show.legend = FALSE, size = 5) +
geom_polygon(data = Compvar_border, aes(fill = Group), color = "black", alpha = 0.3, linetype = 2, show.legend = FALSE) +
scale_color_manual(values = group_colors)
pl4
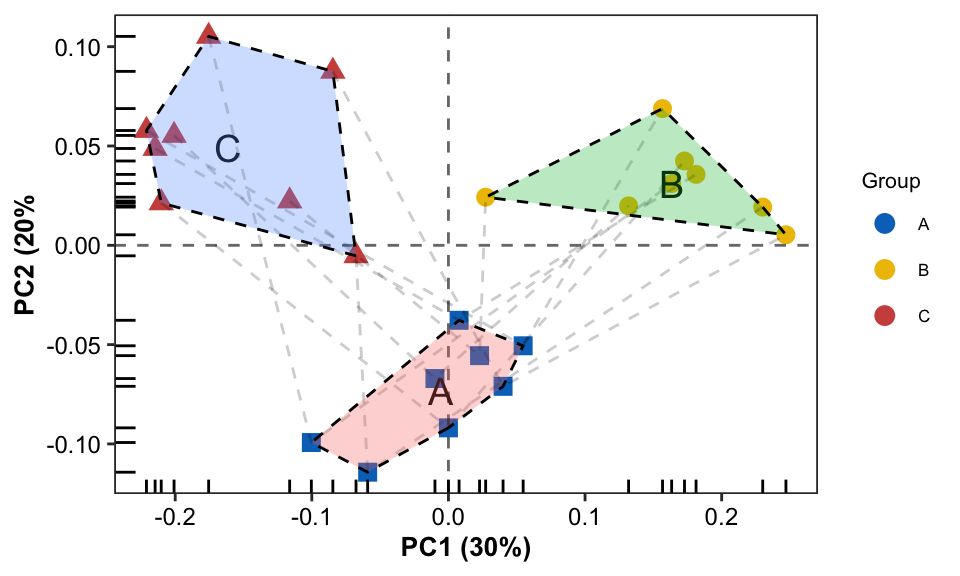
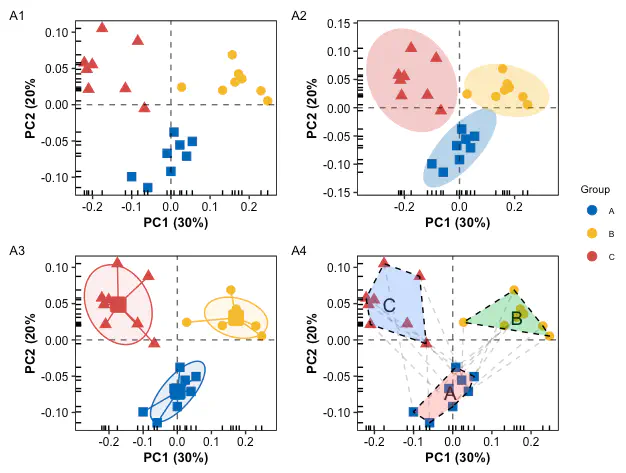
Combination
pl0 + pl1 + pl3 + pl4 + plot_layout(ncol = 2, guides = "collect") +
plot_annotation(tag_levels = c(1), tag_prefix = "A")
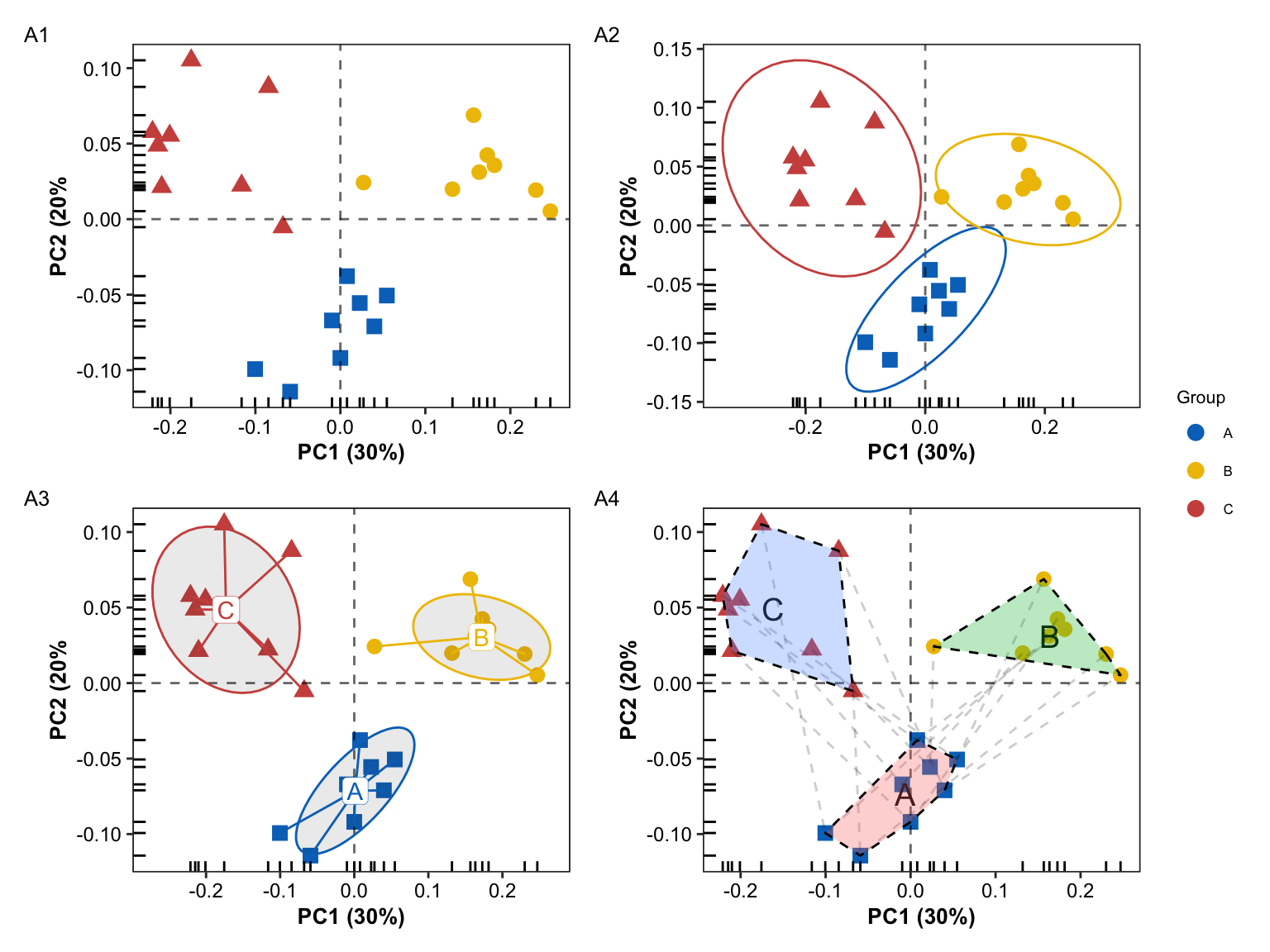
Conclusion
Compared to the single PCA scatterplot, the scatterplot with the addition of the group boundary can more directly reflect the difference information between groups.
Systemic information
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.3 (2022-03-10)
## os macOS Big Sur/Monterey 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2023-08-05
## pandoc 3.1.3 @ /Users/zouhua/opt/anaconda3/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## blogdown 1.18 2023-06-19 [2] CRAN (R 4.1.3)
## bookdown 0.34 2023-05-09 [2] CRAN (R 4.1.2)
## bslib 0.5.0 2023-06-09 [2] CRAN (R 4.1.3)
## cachem 1.0.8 2023-05-01 [2] CRAN (R 4.1.2)
## callr 3.7.3 2022-11-02 [2] CRAN (R 4.1.2)
## cli 3.6.1 2023-03-23 [2] CRAN (R 4.1.2)
## colorspace 2.1-0 2023-01-23 [2] CRAN (R 4.1.2)
## crayon 1.5.2 2022-09-29 [2] CRAN (R 4.1.2)
## devtools 2.4.5 2022-10-11 [2] CRAN (R 4.1.2)
## digest 0.6.33 2023-07-07 [1] CRAN (R 4.1.3)
## dplyr * 1.1.2 2023-04-20 [2] CRAN (R 4.1.2)
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.1.0)
## evaluate 0.21 2023-05-05 [2] CRAN (R 4.1.2)
## fansi 1.0.4 2023-01-22 [2] CRAN (R 4.1.2)
## farver 2.1.1 2022-07-06 [2] CRAN (R 4.1.2)
## fastmap 1.1.1 2023-02-24 [2] CRAN (R 4.1.2)
## forcats * 1.0.0 2023-01-29 [2] CRAN (R 4.1.2)
## fs 1.6.2 2023-04-25 [2] CRAN (R 4.1.2)
## generics 0.1.3 2022-07-05 [2] CRAN (R 4.1.2)
## ggplot2 * 3.4.2 2023-04-03 [2] CRAN (R 4.1.2)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.1.2)
## gtable 0.3.3 2023-03-21 [2] CRAN (R 4.1.2)
## highr 0.10 2022-12-22 [2] CRAN (R 4.1.2)
## hms 1.1.3 2023-03-21 [2] CRAN (R 4.1.2)
## htmltools 0.5.5 2023-03-23 [2] CRAN (R 4.1.2)
## htmlwidgets 1.6.2 2023-03-17 [2] CRAN (R 4.1.2)
## httpuv 1.6.11 2023-05-11 [2] CRAN (R 4.1.3)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.1.0)
## jsonlite 1.8.7 2023-06-29 [2] CRAN (R 4.1.3)
## knitr 1.43 2023-05-25 [2] CRAN (R 4.1.3)
## labeling 0.4.2 2020-10-20 [2] CRAN (R 4.1.0)
## later 1.3.1 2023-05-02 [2] CRAN (R 4.1.2)
## lifecycle 1.0.3 2022-10-07 [2] CRAN (R 4.1.2)
## lubridate * 1.9.2 2023-02-10 [2] CRAN (R 4.1.2)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.1.2)
## MASS 7.3-60 2023-05-04 [2] CRAN (R 4.1.2)
## memoise 2.0.1 2021-11-26 [2] CRAN (R 4.1.0)
## mime 0.12 2021-09-28 [2] CRAN (R 4.1.0)
## miniUI 0.1.1.1 2018-05-18 [2] CRAN (R 4.1.0)
## munsell 0.5.0 2018-06-12 [2] CRAN (R 4.1.0)
## patchwork * 1.1.2 2022-08-19 [2] CRAN (R 4.1.2)
## pillar 1.9.0 2023-03-22 [2] CRAN (R 4.1.2)
## pkgbuild 1.4.2 2023-06-26 [2] CRAN (R 4.1.3)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.1.0)
## pkgload 1.3.2.1 2023-07-08 [2] CRAN (R 4.1.3)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.1.0)
## processx 3.8.2 2023-06-30 [2] CRAN (R 4.1.3)
## profvis 0.3.8 2023-05-02 [2] CRAN (R 4.1.2)
## promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.1.0)
## ps 1.7.5 2023-04-18 [2] CRAN (R 4.1.2)
## purrr * 1.0.1 2023-01-10 [2] CRAN (R 4.1.2)
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.1.0)
## Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.1.3)
## readr * 2.1.4 2023-02-10 [2] CRAN (R 4.1.2)
## remotes 2.4.2 2021-11-30 [2] CRAN (R 4.1.0)
## rlang 1.1.1 2023-04-28 [2] CRAN (R 4.1.2)
## rmarkdown 2.23 2023-07-01 [2] CRAN (R 4.1.3)
## rstudioapi 0.15.0 2023-07-07 [2] CRAN (R 4.1.3)
## sass 0.4.6 2023-05-03 [2] CRAN (R 4.1.2)
## scales 1.2.1 2022-08-20 [2] CRAN (R 4.1.2)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.1.0)
## shiny 1.7.4.1 2023-07-06 [2] CRAN (R 4.1.3)
## stringi 1.7.12 2023-01-11 [2] CRAN (R 4.1.2)
## stringr * 1.5.0 2022-12-02 [2] CRAN (R 4.1.2)
## tibble * 3.2.1 2023-03-20 [2] CRAN (R 4.1.2)
## tidyr * 1.3.0 2023-01-24 [2] CRAN (R 4.1.2)
## tidyselect 1.2.0 2022-10-10 [2] CRAN (R 4.1.2)
## tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.1.2)
## timechange 0.2.0 2023-01-11 [2] CRAN (R 4.1.2)
## tzdb 0.4.0 2023-05-12 [2] CRAN (R 4.1.3)
## urlchecker 1.0.1 2021-11-30 [2] CRAN (R 4.1.0)
## usethis 2.2.2 2023-07-06 [2] CRAN (R 4.1.3)
## utf8 1.2.3 2023-01-31 [2] CRAN (R 4.1.2)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.1.3)
## withr 2.5.0 2022-03-03 [2] CRAN (R 4.1.2)
## xfun 0.39 2023-04-20 [2] CRAN (R 4.1.2)
## xtable 1.8-4 2019-04-21 [2] CRAN (R 4.1.0)
## yaml 2.3.7 2023-01-23 [2] CRAN (R 4.1.2)
##
## [1] /Users/zouhua/Library/R/x86_64/4.1/library
## [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────