R visualization: STAMP by ggplot2
STAMP with barplot and error bar plot
Introduction
STAMP shows the mean abundance/value/proportion of features and the statistical results (t-test or wilcox-test) between groups.
Loading required packages
knitr::opts_chunk$set(message = FALSE, warning = FALSE)
library(tidyverse)
library(broom)
library(patchwork)
rm(list = ls())
options(stringsAsFactors = F)
group_names <- c("setosa", "versicolor")
group_colors <- c("#E69F00", "#56B4E9")
Data preparation for hypothesis testing
Loading iris dataset
Factorizing Species
data("iris")
test_data <- iris |>
dplyr::select(Species, Sepal.Length, Sepal.Width, Petal.Length, Petal.Width) |>
dplyr::filter(Species %in% group_names) |>
dplyr::mutate(Species = factor(Species, levels = group_names))
head(test_data)
## Species Sepal.Length Sepal.Width Petal.Length Petal.Width
## 1 setosa 5.1 3.5 1.4 0.2
## 2 setosa 4.9 3.0 1.4 0.2
## 3 setosa 4.7 3.2 1.3 0.2
## 4 setosa 4.6 3.1 1.5 0.2
## 5 setosa 5.0 3.6 1.4 0.2
## 6 setosa 5.4 3.9 1.7 0.4
- T-test for normal distribution, otherwise wilcox’s test
test_res <- test_data %>%
dplyr::select_if(is.numeric) %>%
purrr::map_df(~ broom::tidy(t.test(. ~ Species, data = test_data)), .id = 'Vars') |>
dplyr::mutate(adjustP = p.adjust(p.value, method = "BH")) |>
dplyr::filter(adjustP < 0.05)
# test_res <- test_data %>%
# dplyr::select_if(is.numeric) %>%
# purrr::map_df(~ broom::tidy(wilcox.test(. ~ Species, data = test_data)), .id = 'Vars') |>
# dplyr::mutate(adjustP = p.adjust(p.value, method = "BH")) |>
# dplyr::filter(adjustP < 0.05)
head(test_res)
## # A tibble: 4 × 12
## Vars estimate estimate1 estimate2 statistic p.value parameter conf.low
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Sepal.Leng… -0.93 5.01 5.94 -10.5 3.75e-17 86.5 -1.11
## 2 Sepal.Width 0.658 3.43 2.77 9.45 2.48e-15 94.7 0.520
## 3 Petal.Leng… -2.80 1.46 4.26 -39.5 9.93e-46 62.1 -2.94
## 4 Petal.Width -1.08 0.246 1.33 -34.1 2.72e-47 74.8 -1.14
## # ℹ 4 more variables: conf.high <dbl>, method <chr>, alternative <chr>,
## # adjustP <dbl>
Data preparation for plotting
- data for left subfigure, which is barplot
data_left <- test_data |>
dplyr::select(all_of(c("Species", test_res$Vars))) |>
tidyr::pivot_longer(cols = -Species,
names_to = "Variables",
values_to = "Values") |>
dplyr::group_by(Variables, Species) %>%
dplyr::summarise(Mean = mean(Values), .groups = "drop")
head(data_left)
## # A tibble: 6 × 3
## Variables Species Mean
## <chr> <fct> <dbl>
## 1 Petal.Length setosa 1.46
## 2 Petal.Length versicolor 4.26
## 3 Petal.Width setosa 0.246
## 4 Petal.Width versicolor 1.33
## 5 Sepal.Length setosa 5.01
## 6 Sepal.Length versicolor 5.94
- data for right subfigure, which is error bar plot
data_right <- test_res |>
dplyr::select(Vars, estimate, conf.low, conf.high, p.value, adjustP) |>
dplyr::mutate(Species = ifelse(estimate > 0, group_names[1], group_names[2])) |>
dplyr::arrange(desc(estimate)) |>
dplyr::mutate(p.value = signif(p.value, 3),
adjustP = signif(adjustP, 3)) |>
dplyr::mutate(p.value = as.character(p.value),
adjustP = as.character(adjustP))
head(data_right)
## # A tibble: 4 × 7
## Vars estimate conf.low conf.high p.value adjustP Species
## <chr> <dbl> <dbl> <dbl> <chr> <chr> <chr>
## 1 Sepal.Width 0.658 0.520 0.796 2.48e-15 2.48e-15 setosa
## 2 Sepal.Length -0.93 -1.11 -0.754 3.75e-17 5e-17 versicolor
## 3 Petal.Width -1.08 -1.14 -1.02 2.72e-47 1.09e-46 versicolor
## 4 Petal.Length -2.80 -2.94 -2.66 9.93e-46 1.99e-45 versicolor
- Order variables in two dataset
data_left$Variables <- factor(data_left$Variables, levels = rev(data_right$Vars))
data_right$Vars <- factor(data_right$Vars, levels = levels(data_left$Variables))
Plotting
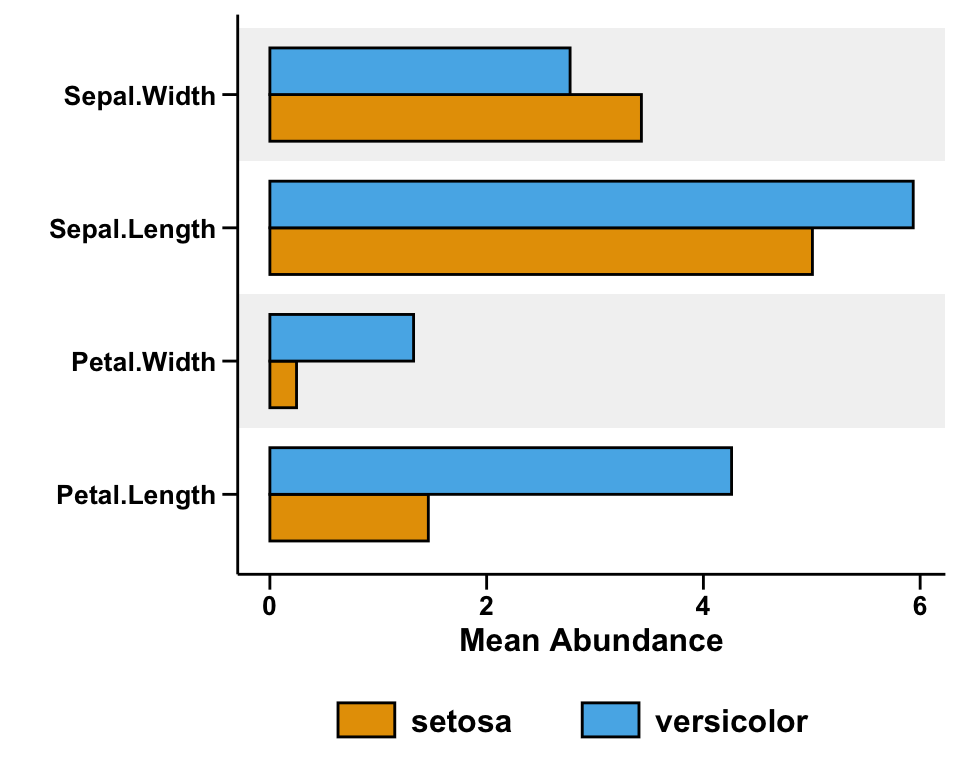
left subfigure
apply
geom_barfor barplotadd segement by
annotate
pl_left_bar <- ggplot(data_left, aes(x = Variables, y = Mean, fill = Species)) +
scale_x_discrete(limits = levels(data_left$Variables)) +
coord_flip() +
labs(x = "", y = "Mean Abundance") +
theme(panel.background = element_rect(fill = "transparent"),
panel.grid = element_blank(),
axis.ticks.length = unit(0.4, "lines"),
axis.ticks = element_line(color = "black"),
axis.line = element_line(color = "black"),
axis.title.x = element_text(color = "black", size = 12, face = "bold"),
axis.text = element_text(color = "black", size = 10, face = "bold"),
legend.title = element_blank(),
legend.text = element_text(size = 12, face = "bold",
color = "black", margin = margin(r = 20)),
legend.position = "bottom",
legend.direction = "horizontal",
legend.key.width = unit(0.8, "cm"),
legend.key.height = unit(0.5, "cm"))
for (i in 1:(nrow(data_right) - 1)) {
pl_left_bar <- pl_left_bar + annotate("rect", xmin = i + 0.5, xmax = i+1.5, ymin = -Inf, ymax = Inf,
fill = ifelse(i %% 2 == 0, 'white', 'gray95'))
}
pl_left_bar <- pl_left_bar +
geom_bar(stat = "identity", position = "dodge", width = 0.7, color = "black") +
scale_fill_manual(values = group_colors)
pl_left_bar
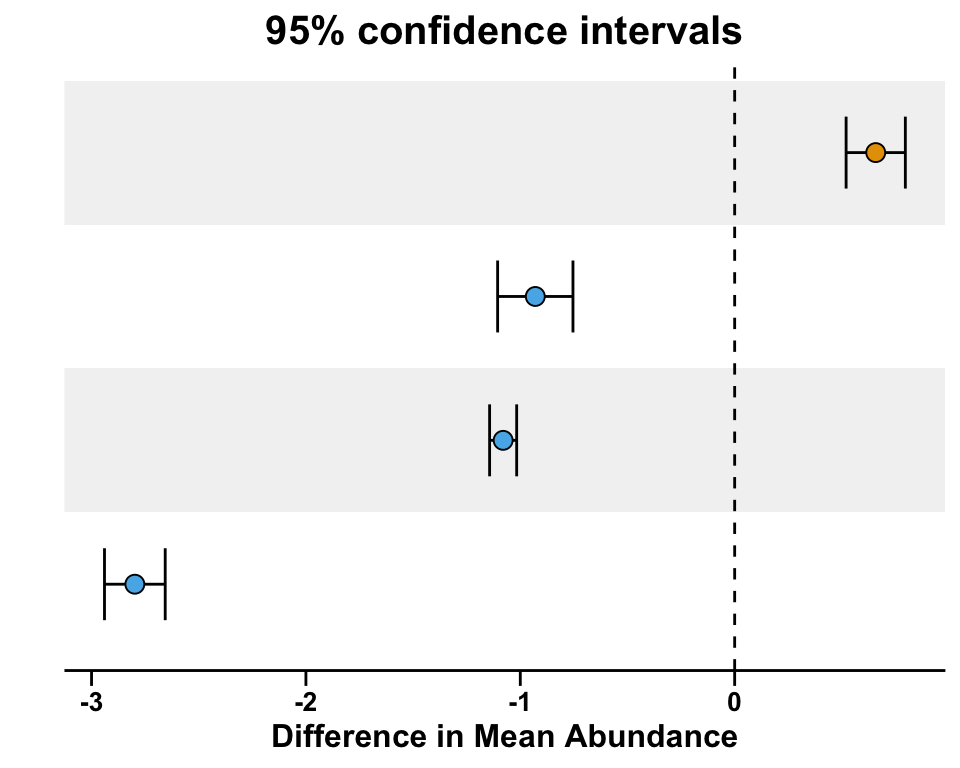
right subfigure for error bar
apply
geom_errorbarfor error baradd segement by
annotate
pl_right_scatter <- ggplot(data_right, aes(x = Vars, y = estimate, fill = Species)) +
theme(panel.background = element_rect(fill = 'transparent'),
panel.grid = element_blank(),
axis.ticks.length = unit(0.4, "lines"),
axis.ticks = element_line(color = "black"),
axis.line = element_line(color = "black"),
axis.title.x = element_text(color = "black", size = 12,face = "bold"),
axis.text = element_text(color = "black", size = 10, face = "bold"),
axis.text.y = element_blank(),
legend.position = "none",
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
plot.title = element_text(size = 15, face = "bold", color = "black", hjust = 0.5)) +
scale_x_discrete(limits = levels(data_right$Vars)) +
coord_flip() +
xlab("") +
ylab("Difference in Mean Abundance") +
labs(title="95% confidence intervals")
for (i in 1:(nrow(data_right) - 1)) {
pl_right_scatter <- pl_right_scatter +
annotate("rect", xmin = i+0.5, xmax = i+1.5, ymin = -Inf, ymax = Inf,
fill = ifelse(i %% 2 == 0, "white", "gray95"))
}
pl_right_scatter <- pl_right_scatter +
geom_errorbar(aes(ymin = conf.low, ymax = conf.high),
position = position_dodge(0.8), width = 0.5, linewidth = 0.5) +
geom_point(shape = 21, size = 3) +
scale_fill_manual(values = group_colors) +
geom_hline(aes(yintercept = 0), linetype = 'dashed', color = 'black')
pl_right_scatter
- pvalue subfigure
pl_right_pvalue <- ggplot(data_right, aes(x = Vars, y = estimate, fill = Species)) +
geom_text(aes(y = 0, x = Vars), label = data_right$adjustP,
hjust = 0, fontface = "bold", inherit.aes = FALSE, size = 3) +
geom_text(aes(x = nrow(data_right)/2 + 0.5, y = 0.85), label = "Adjusted-Pvalue",
srt = 90, fontface = "bold", size = 3) +
coord_flip() +
ylim(c(0, 1)) +
theme(panel.background = element_blank(),
panel.grid = element_blank(),
axis.line = element_blank(),
axis.ticks = element_blank(),
axis.text = element_blank(),
axis.title = element_blank())
pl_right_pvalue

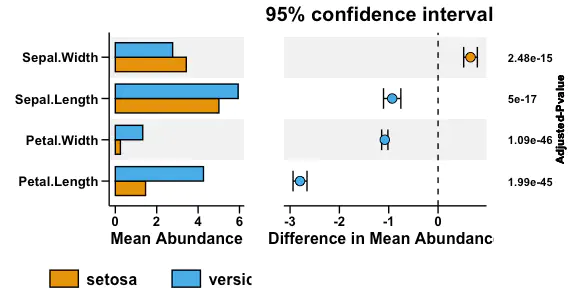
- Combination of all subfigures
pl <- pl_left_bar + pl_right_scatter + pl_right_pvalue +
plot_layout(widths = c(4, 6, 2))
pl
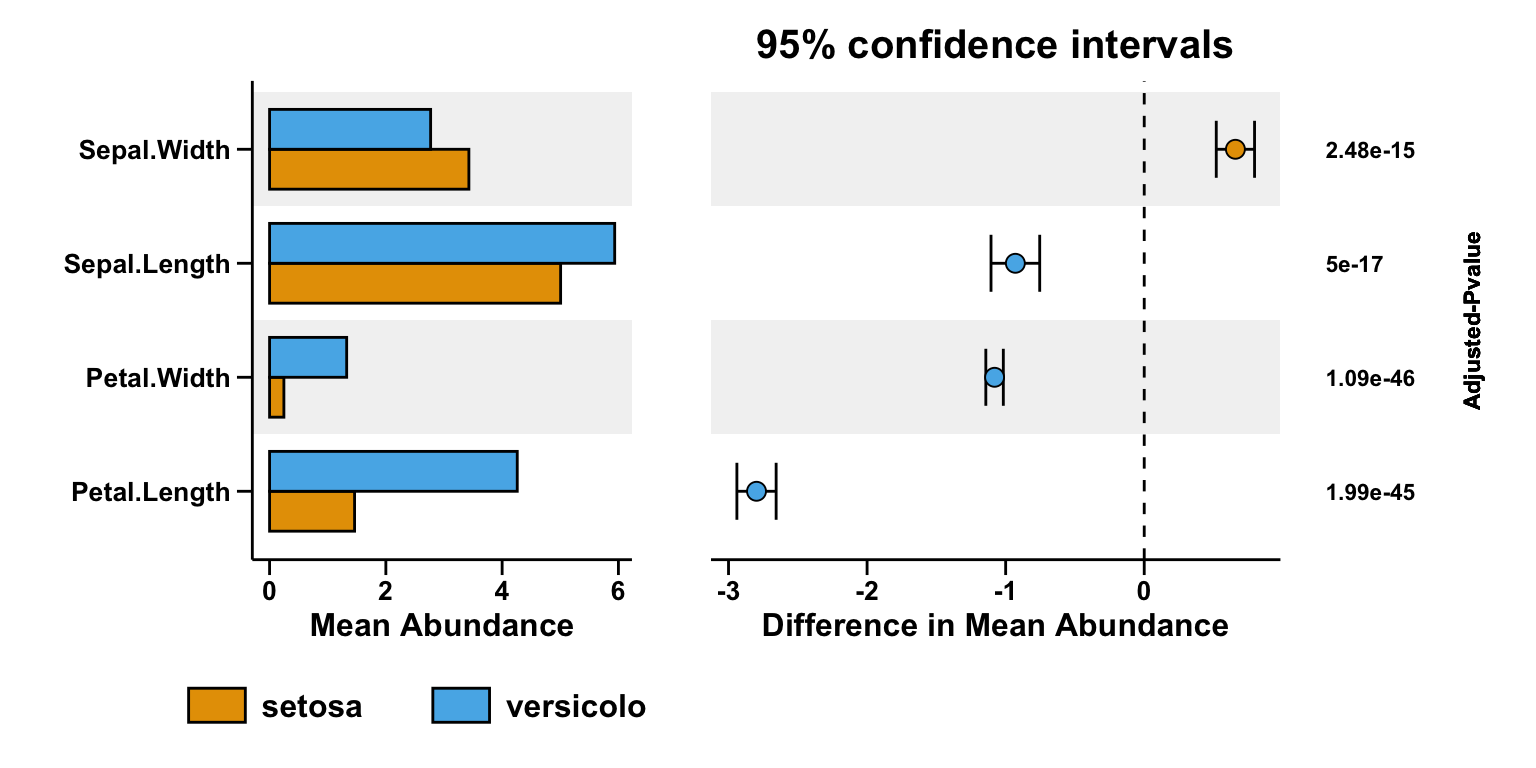
Conclusion
STAMP is comprise of barplot for mean abundance and error bar plot for hypothesis testing.
Systemic information
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.3 (2022-03-10)
## os macOS Big Sur/Monterey 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2023-07-24
## pandoc 3.1.3 @ /Users/zouhua/opt/anaconda3/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## backports 1.4.1 2021-12-13 [2] CRAN (R 4.1.0)
## blogdown 1.18 2023-06-19 [2] CRAN (R 4.1.3)
## bookdown 0.34 2023-05-09 [2] CRAN (R 4.1.2)
## broom * 1.0.5 2023-06-09 [2] CRAN (R 4.1.3)
## bslib 0.5.0 2023-06-09 [2] CRAN (R 4.1.3)
## cachem 1.0.8 2023-05-01 [2] CRAN (R 4.1.2)
## callr 3.7.3 2022-11-02 [2] CRAN (R 4.1.2)
## cli 3.6.1 2023-03-23 [2] CRAN (R 4.1.2)
## colorspace 2.1-0 2023-01-23 [2] CRAN (R 4.1.2)
## crayon 1.5.2 2022-09-29 [2] CRAN (R 4.1.2)
## devtools 2.4.5 2022-10-11 [2] CRAN (R 4.1.2)
## digest 0.6.33 2023-07-07 [1] CRAN (R 4.1.3)
## dplyr * 1.1.2 2023-04-20 [2] CRAN (R 4.1.2)
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.1.0)
## evaluate 0.21 2023-05-05 [2] CRAN (R 4.1.2)
## fansi 1.0.4 2023-01-22 [2] CRAN (R 4.1.2)
## farver 2.1.1 2022-07-06 [2] CRAN (R 4.1.2)
## fastmap 1.1.1 2023-02-24 [2] CRAN (R 4.1.2)
## forcats * 1.0.0 2023-01-29 [2] CRAN (R 4.1.2)
## fs 1.6.2 2023-04-25 [2] CRAN (R 4.1.2)
## generics 0.1.3 2022-07-05 [2] CRAN (R 4.1.2)
## ggplot2 * 3.4.2 2023-04-03 [2] CRAN (R 4.1.2)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.1.2)
## gtable 0.3.3 2023-03-21 [2] CRAN (R 4.1.2)
## highr 0.10 2022-12-22 [2] CRAN (R 4.1.2)
## hms 1.1.3 2023-03-21 [2] CRAN (R 4.1.2)
## htmltools 0.5.5 2023-03-23 [2] CRAN (R 4.1.2)
## htmlwidgets 1.6.2 2023-03-17 [2] CRAN (R 4.1.2)
## httpuv 1.6.11 2023-05-11 [2] CRAN (R 4.1.3)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.1.0)
## jsonlite 1.8.7 2023-06-29 [2] CRAN (R 4.1.3)
## knitr 1.43 2023-05-25 [2] CRAN (R 4.1.3)
## labeling 0.4.2 2020-10-20 [2] CRAN (R 4.1.0)
## later 1.3.1 2023-05-02 [2] CRAN (R 4.1.2)
## lifecycle 1.0.3 2022-10-07 [2] CRAN (R 4.1.2)
## lubridate * 1.9.2 2023-02-10 [2] CRAN (R 4.1.2)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.1.2)
## memoise 2.0.1 2021-11-26 [2] CRAN (R 4.1.0)
## mime 0.12 2021-09-28 [2] CRAN (R 4.1.0)
## miniUI 0.1.1.1 2018-05-18 [2] CRAN (R 4.1.0)
## munsell 0.5.0 2018-06-12 [2] CRAN (R 4.1.0)
## patchwork * 1.1.2 2022-08-19 [2] CRAN (R 4.1.2)
## pillar 1.9.0 2023-03-22 [2] CRAN (R 4.1.2)
## pkgbuild 1.4.2 2023-06-26 [2] CRAN (R 4.1.3)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.1.0)
## pkgload 1.3.2.1 2023-07-08 [2] CRAN (R 4.1.3)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.1.0)
## processx 3.8.2 2023-06-30 [2] CRAN (R 4.1.3)
## profvis 0.3.8 2023-05-02 [2] CRAN (R 4.1.2)
## promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.1.0)
## ps 1.7.5 2023-04-18 [2] CRAN (R 4.1.2)
## purrr * 1.0.1 2023-01-10 [2] CRAN (R 4.1.2)
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.1.0)
## Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.1.3)
## readr * 2.1.4 2023-02-10 [2] CRAN (R 4.1.2)
## remotes 2.4.2 2021-11-30 [2] CRAN (R 4.1.0)
## rlang 1.1.1 2023-04-28 [2] CRAN (R 4.1.2)
## rmarkdown 2.23 2023-07-01 [2] CRAN (R 4.1.3)
## rstudioapi 0.15.0 2023-07-07 [2] CRAN (R 4.1.3)
## sass 0.4.6 2023-05-03 [2] CRAN (R 4.1.2)
## scales 1.2.1 2022-08-20 [2] CRAN (R 4.1.2)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.1.0)
## shiny 1.7.4.1 2023-07-06 [2] CRAN (R 4.1.3)
## stringi 1.7.12 2023-01-11 [2] CRAN (R 4.1.2)
## stringr * 1.5.0 2022-12-02 [2] CRAN (R 4.1.2)
## tibble * 3.2.1 2023-03-20 [2] CRAN (R 4.1.2)
## tidyr * 1.3.0 2023-01-24 [2] CRAN (R 4.1.2)
## tidyselect 1.2.0 2022-10-10 [2] CRAN (R 4.1.2)
## tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.1.2)
## timechange 0.2.0 2023-01-11 [2] CRAN (R 4.1.2)
## tzdb 0.4.0 2023-05-12 [2] CRAN (R 4.1.3)
## urlchecker 1.0.1 2021-11-30 [2] CRAN (R 4.1.0)
## usethis 2.2.2 2023-07-06 [2] CRAN (R 4.1.3)
## utf8 1.2.3 2023-01-31 [2] CRAN (R 4.1.2)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.1.3)
## withr 2.5.0 2022-03-03 [2] CRAN (R 4.1.2)
## xfun 0.39 2023-04-20 [2] CRAN (R 4.1.2)
## xtable 1.8-4 2019-04-21 [2] CRAN (R 4.1.0)
## yaml 2.3.7 2023-01-23 [2] CRAN (R 4.1.2)
##
## [1] /Users/zouhua/Library/R/x86_64/4.1/library
## [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────