R visualization: Violin by ggplot2
Violin with boxplot and point
Introduction
Violin using the density of distribution to show the differences among groups is one of the most important means for data visualization.
Loading required packages
knitr::opts_chunk$set(message = FALSE, warning = FALSE)
library(tidyverse)
library(ggpubr)
# install.packages("gghalves")
library(gghalves)
rm(list = ls())
options(stringsAsFactors = F)
# group & color
group_names <- c("setosa", "versicolor", "virginica")
group_colors <- c("#0073C2FF", "#EFC000FF", "#CD534CFF")
Data preparation
Loading iris and ToothGrowth dataset
Factorizing Species
data("iris")
plotdata <- iris |>
dplyr::select(Sepal.Length, Species, Sepal.Width) |>
dplyr::mutate(Species = factor(Species, levels = group_names)) |>
dplyr::rename(Group = Species,
Index = Sepal.Length,
Index2 = Sepal.Width)
head(plotdata)
## Index Group Index2
## 1 5.1 setosa 3.5
## 2 4.9 setosa 3.0
## 3 4.7 setosa 3.2
## 4 4.6 setosa 3.1
## 5 5.0 setosa 3.6
## 6 5.4 setosa 3.9
# other dataset
data("ToothGrowth")
Single violin
create violin by
geom_violinapply
stat_boxplotandgeom_boxplotfor boxplotadd point by
geom_point
pl_violin <- ggplot(data = plotdata, aes(x = Group, y = Index, shape = Group)) +
geom_violin(aes(fill = Group), alpha = 0.5) +
stat_boxplot(geom = "errorbar", width = 0.2) +
geom_boxplot(width = .1, notch = TRUE) +
geom_point(aes(color = Group), position = position_jitter(width = 0.1), size = 1) +
stat_compare_means(comparisons = list(c("setosa", "versicolor"),
c("setosa", "virginica")),
method = "t.test",
label = "p.signif") +
labs(x = "") +
scale_y_continuous(expand = expansion(mult = c(0.1, 0.1))) +
scale_color_manual(values = group_colors) +
scale_fill_manual(values = group_colors) +
scale_shape_manual(values = c(15, 16, 17)) +
guides(color = "none", shape = "none", fill = "none") +
theme_bw() +
theme(axis.title = element_text(size = 12, color = "black", face = "bold"),
axis.text = element_text(size = 10, color = "black"),
text = element_text(size = 9, color = "black"))
pl_violin
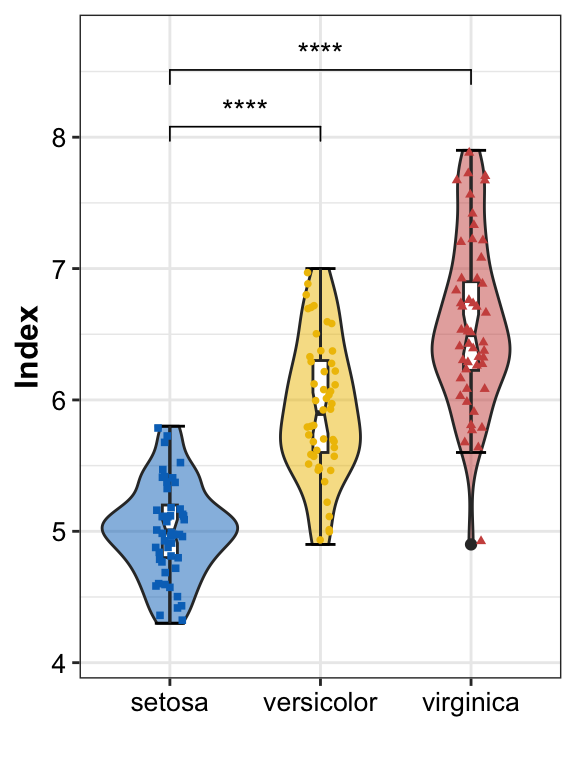
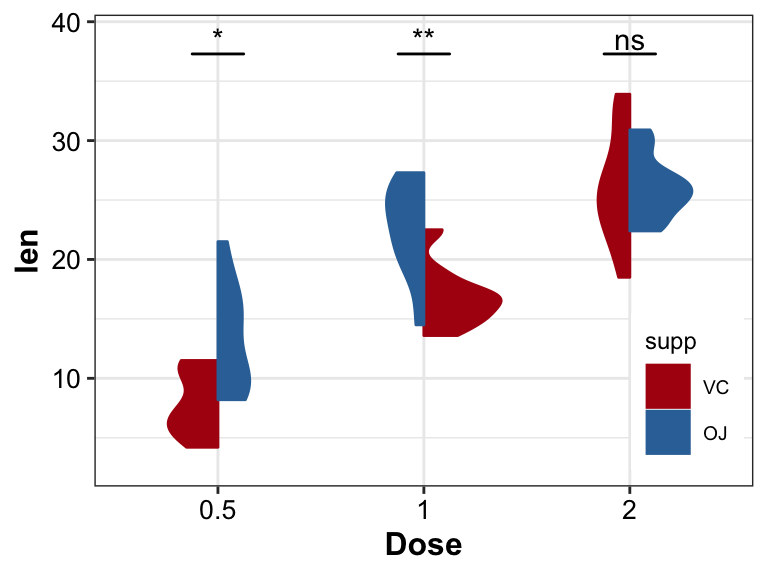
Violin with two different groups
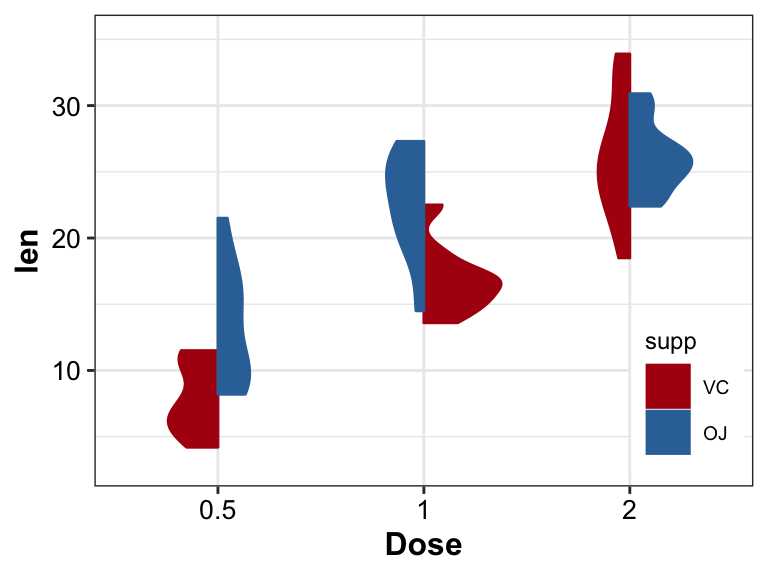
The left part of violin is totally different from the right.
- Using ToothGrowth dataset with groups Dose and supp
pl_violin2 <- ToothGrowth |>
dplyr::mutate(supp = factor(supp, levels = c("VC", "OJ"))) |>
ggplot(aes(x = as.factor(dose), y = len)) +
geom_half_violin(aes(fill = supp, color = supp, split = supp),
position = "identity") +
labs(x = "Dose") +
scale_y_continuous(expand = expansion(mult = c(0.1, 0.1))) +
scale_color_manual(values = c("#AF0F11", "#3372A6")) +
scale_fill_manual(values = c("#AF0F11", "#3372A6")) +
theme_bw() +
theme(axis.title = element_text(size = 12, color = "black", face = "bold"),
axis.text = element_text(size = 10, color = "black"),
text = element_text(size = 9, color = "black"),
legend.position = c(.9, 0.2))
pl_violin2
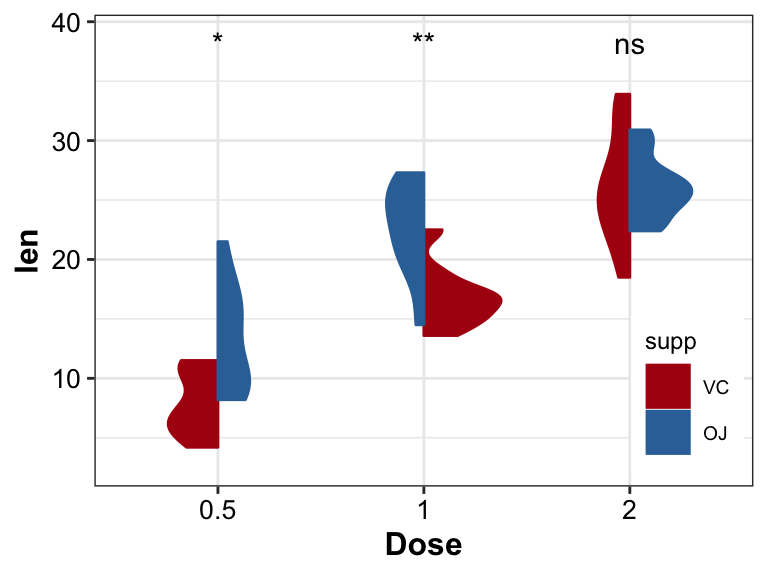
- add significance between groups by
stat_compare_means
pl_violin2_1 <- pl_violin2 +
stat_compare_means(aes(group = supp),
label = "p.signif",
label.y = (max(ToothGrowth$len) + max(ToothGrowth$len) * 0.1))
pl_violin2_1
- add significance between groups by
ggsignif::geom_signif
library(ggsignif)
nlength <- length(unique(ToothGrowth$dose))
test_res <- ToothGrowth |>
dplyr::mutate(dose = factor(dose)) |>
dplyr::group_by(dose) |>
rstatix::t_test(len ~ supp) |>
rstatix::adjust_pvalue() |>
rstatix::add_significance("p.adj") |>
dplyr::mutate(x = seq(0.875, 0.875+(nlength-1)*1, 1),
xend = seq(1.125, 1.125+(nlength-1)*1, 1),
y = rep((max(ToothGrowth$len) + max(ToothGrowth$len) * 0.1), nlength))
pl_violin2_2 <- pl_violin2 +
geom_signif(stat = "identity",
data = test_res,
aes(x = x,xend = xend, y = y, yend = y, annotation = p.adj.signif))
pl_violin2_2
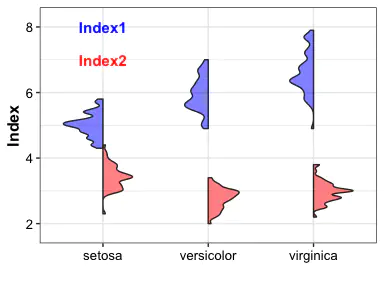
Violin with two different index
pl_violin3 <- ggplot(data = plotdata, aes(x = Group, y = Index)) +
geom_half_violin(fill = "blue",
side = "l",
adjust = 0.5,
alpha = 0.5) +
geom_half_violin(aes(y = Index2),
fill = "red",
side = "r",
adjust = 0.5,
alpha = 0.5) +
labs(x = "") +
annotate(x = 0.5, y = 8, geom = "text", label = "Index", color = "blue",
size = 4, fontface = "bold", hjust = 0) +
annotate(x = 0.5, y = 7.5, geom = "text", label = "Index2", color = "red",
size = 4, fontface = "bold", hjust = 0) +
scale_y_continuous(expand = expansion(mult = c(0.1, 0.1))) +
theme_bw() +
theme(axis.title = element_text(size = 12, color = "black", face = "bold"),
axis.text = element_text(size = 10, color = "black"),
text = element_text(size = 9, color = "black"))
pl_violin3
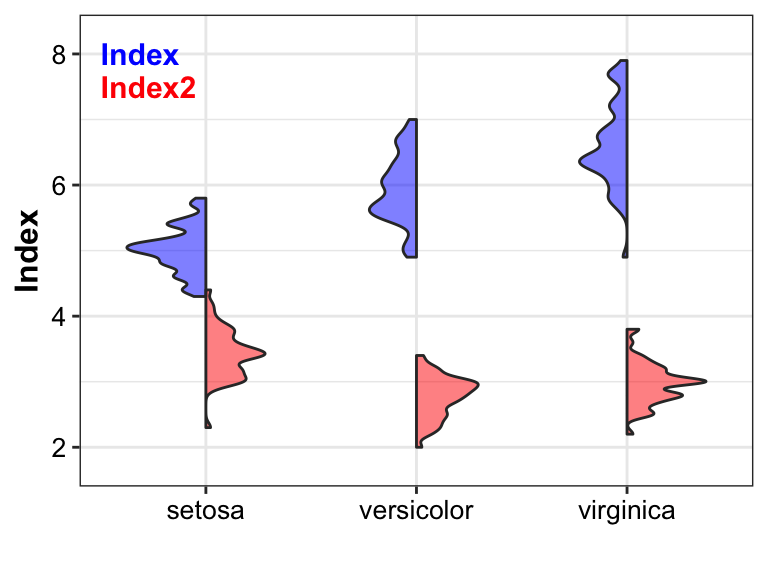
Conclusion
Compared to single boxplot, violin not only shows differences among group, but also displays the distribution of each value, providing more information.
Systemic information
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.3 (2022-03-10)
## os macOS Big Sur/Monterey 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2023-07-24
## pandoc 3.1.3 @ /Users/zouhua/opt/anaconda3/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## abind 1.4-5 2016-07-21 [2] CRAN (R 4.1.0)
## backports 1.4.1 2021-12-13 [2] CRAN (R 4.1.0)
## blogdown 1.18 2023-06-19 [2] CRAN (R 4.1.3)
## bookdown 0.34 2023-05-09 [2] CRAN (R 4.1.2)
## broom 1.0.5 2023-06-09 [2] CRAN (R 4.1.3)
## bslib 0.5.0 2023-06-09 [2] CRAN (R 4.1.3)
## cachem 1.0.8 2023-05-01 [2] CRAN (R 4.1.2)
## callr 3.7.3 2022-11-02 [2] CRAN (R 4.1.2)
## car 3.1-2 2023-03-30 [2] CRAN (R 4.1.2)
## carData 3.0-5 2022-01-06 [2] CRAN (R 4.1.2)
## cli 3.6.1 2023-03-23 [2] CRAN (R 4.1.2)
## colorspace 2.1-0 2023-01-23 [2] CRAN (R 4.1.2)
## crayon 1.5.2 2022-09-29 [2] CRAN (R 4.1.2)
## devtools 2.4.5 2022-10-11 [2] CRAN (R 4.1.2)
## digest 0.6.33 2023-07-07 [1] CRAN (R 4.1.3)
## dplyr * 1.1.2 2023-04-20 [2] CRAN (R 4.1.2)
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.1.0)
## evaluate 0.21 2023-05-05 [2] CRAN (R 4.1.2)
## fansi 1.0.4 2023-01-22 [2] CRAN (R 4.1.2)
## farver 2.1.1 2022-07-06 [2] CRAN (R 4.1.2)
## fastmap 1.1.1 2023-02-24 [2] CRAN (R 4.1.2)
## forcats * 1.0.0 2023-01-29 [2] CRAN (R 4.1.2)
## fs 1.6.2 2023-04-25 [2] CRAN (R 4.1.2)
## generics 0.1.3 2022-07-05 [2] CRAN (R 4.1.2)
## gghalves * 0.1.4 2022-11-20 [1] CRAN (R 4.1.2)
## ggplot2 * 3.4.2 2023-04-03 [2] CRAN (R 4.1.2)
## ggpubr * 0.6.0 2023-02-10 [2] CRAN (R 4.1.2)
## ggsignif * 0.6.4 2022-10-13 [2] CRAN (R 4.1.2)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.1.2)
## gtable 0.3.3 2023-03-21 [2] CRAN (R 4.1.2)
## highr 0.10 2022-12-22 [2] CRAN (R 4.1.2)
## hms 1.1.3 2023-03-21 [2] CRAN (R 4.1.2)
## htmltools 0.5.5 2023-03-23 [2] CRAN (R 4.1.2)
## htmlwidgets 1.6.2 2023-03-17 [2] CRAN (R 4.1.2)
## httpuv 1.6.11 2023-05-11 [2] CRAN (R 4.1.3)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.1.0)
## jsonlite 1.8.7 2023-06-29 [2] CRAN (R 4.1.3)
## knitr 1.43 2023-05-25 [2] CRAN (R 4.1.3)
## labeling 0.4.2 2020-10-20 [2] CRAN (R 4.1.0)
## later 1.3.1 2023-05-02 [2] CRAN (R 4.1.2)
## lifecycle 1.0.3 2022-10-07 [2] CRAN (R 4.1.2)
## lubridate * 1.9.2 2023-02-10 [2] CRAN (R 4.1.2)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.1.2)
## memoise 2.0.1 2021-11-26 [2] CRAN (R 4.1.0)
## mime 0.12 2021-09-28 [2] CRAN (R 4.1.0)
## miniUI 0.1.1.1 2018-05-18 [2] CRAN (R 4.1.0)
## munsell 0.5.0 2018-06-12 [2] CRAN (R 4.1.0)
## pillar 1.9.0 2023-03-22 [2] CRAN (R 4.1.2)
## pkgbuild 1.4.2 2023-06-26 [2] CRAN (R 4.1.3)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.1.0)
## pkgload 1.3.2.1 2023-07-08 [2] CRAN (R 4.1.3)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.1.0)
## processx 3.8.2 2023-06-30 [2] CRAN (R 4.1.3)
## profvis 0.3.8 2023-05-02 [2] CRAN (R 4.1.2)
## promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.1.0)
## ps 1.7.5 2023-04-18 [2] CRAN (R 4.1.2)
## purrr * 1.0.1 2023-01-10 [2] CRAN (R 4.1.2)
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.1.0)
## Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.1.3)
## readr * 2.1.4 2023-02-10 [2] CRAN (R 4.1.2)
## remotes 2.4.2 2021-11-30 [2] CRAN (R 4.1.0)
## rlang 1.1.1 2023-04-28 [2] CRAN (R 4.1.2)
## rmarkdown 2.23 2023-07-01 [2] CRAN (R 4.1.3)
## rstatix 0.7.2 2023-02-01 [2] CRAN (R 4.1.2)
## rstudioapi 0.15.0 2023-07-07 [2] CRAN (R 4.1.3)
## sass 0.4.6 2023-05-03 [2] CRAN (R 4.1.2)
## scales 1.2.1 2022-08-20 [2] CRAN (R 4.1.2)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.1.0)
## shiny 1.7.4.1 2023-07-06 [2] CRAN (R 4.1.3)
## stringi 1.7.12 2023-01-11 [2] CRAN (R 4.1.2)
## stringr * 1.5.0 2022-12-02 [2] CRAN (R 4.1.2)
## tibble * 3.2.1 2023-03-20 [2] CRAN (R 4.1.2)
## tidyr * 1.3.0 2023-01-24 [2] CRAN (R 4.1.2)
## tidyselect 1.2.0 2022-10-10 [2] CRAN (R 4.1.2)
## tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.1.2)
## timechange 0.2.0 2023-01-11 [2] CRAN (R 4.1.2)
## tzdb 0.4.0 2023-05-12 [2] CRAN (R 4.1.3)
## urlchecker 1.0.1 2021-11-30 [2] CRAN (R 4.1.0)
## usethis 2.2.2 2023-07-06 [2] CRAN (R 4.1.3)
## utf8 1.2.3 2023-01-31 [2] CRAN (R 4.1.2)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.1.3)
## withr 2.5.0 2022-03-03 [2] CRAN (R 4.1.2)
## xfun 0.39 2023-04-20 [2] CRAN (R 4.1.2)
## xtable 1.8-4 2019-04-21 [2] CRAN (R 4.1.0)
## yaml 2.3.7 2023-01-23 [2] CRAN (R 4.1.2)
##
## [1] /Users/zouhua/Library/R/x86_64/4.1/library
## [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────