R visualization: donut chart by ggplot2
donut chart shows the number or proportion per group
Introduction
Compared to pie chart, donut chart has one more hole inside. In addition, multiple circles plot shows different traits of dataset.
Loading required packages
knitr::opts_chunk$set(message = FALSE, warning = FALSE)
library(ggplot2)
library(dplyr)
library(tidyverse)
# group & color
group_names <- c("setosa", "versicolor", "virginica")
group_colors <- c("#0073C2FF", "#EFC000FF", "#CD534CFF")
Data preparation
Loading iris dataset
Summarizing the total number per species
Calculating proportion per species
Computing position of text label
data("iris")
plotdata <- iris |>
dplyr::group_by(Species) |>
dplyr::summarise(count = length(Species)) |>
dplyr::mutate(prop = round(count / sum(count), 2)) |>
dplyr::arrange(desc(Species)) |>
dplyr::mutate(lab.ypos = cumsum(prop) - 0.5 * prop) |>
dplyr::mutate(num_prop = paste0("n = ", count, "\n",
paste0(prop, "%"))) |>
dplyr::mutate(Species = factor(Species, levels = group_names))
head(plotdata)
## # A tibble: 3 × 5
## Species count prop lab.ypos num_prop
## <fct> <int> <dbl> <dbl> <chr>
## 1 virginica 50 0.33 0.165 "n = 50\n0.33%"
## 2 versicolor 50 0.33 0.495 "n = 50\n0.33%"
## 3 setosa 50 0.33 0.825 "n = 50\n0.33%"
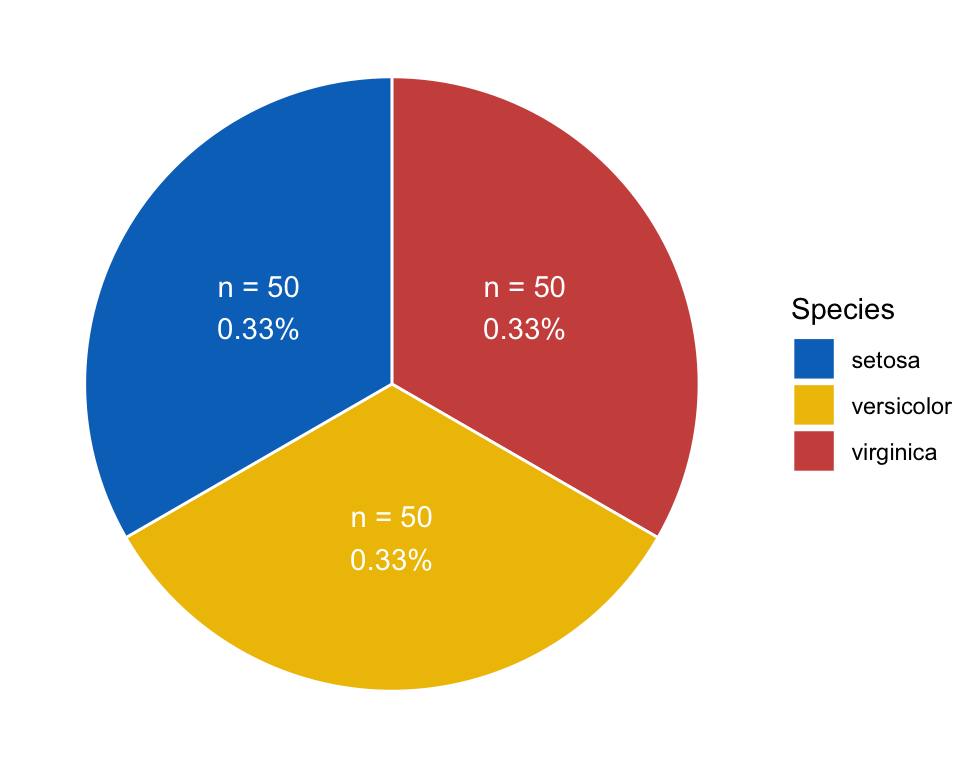
Pie
pie <- ggplot(plotdata, aes(x = "", y = prop, fill = Species)) +
geom_bar(stat = "identity", color = "white", width = 1) +
coord_polar(theta = "y", start = 0) +
geom_text(aes(y = lab.ypos, label = num_prop), color = "white") +
scale_fill_manual(breaks = group_names,
values = group_colors) +
theme_void()
pie
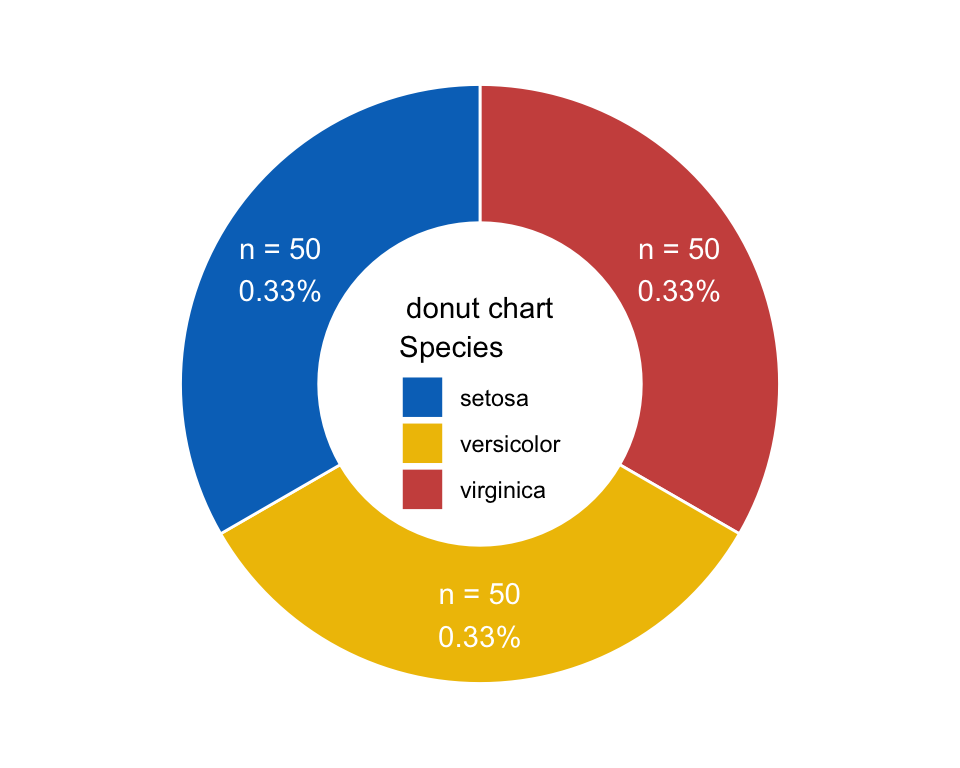
Donut
donut <- ggplot(plotdata, aes(x = 2, y = prop, fill = Species)) +
geom_bar(stat = "identity", color = "white") +
coord_polar(theta = "y", start = 0) +
geom_text(aes(y = lab.ypos, label = num_prop), color = "white") +
scale_fill_manual(breaks = group_names,
values = group_colors) +
annotate("text", x = 1, y = 0, label = "donut chart") +
theme_void() +
xlim(0.5, 2.5) +
theme(legend.position = c(.5, .45))
donut
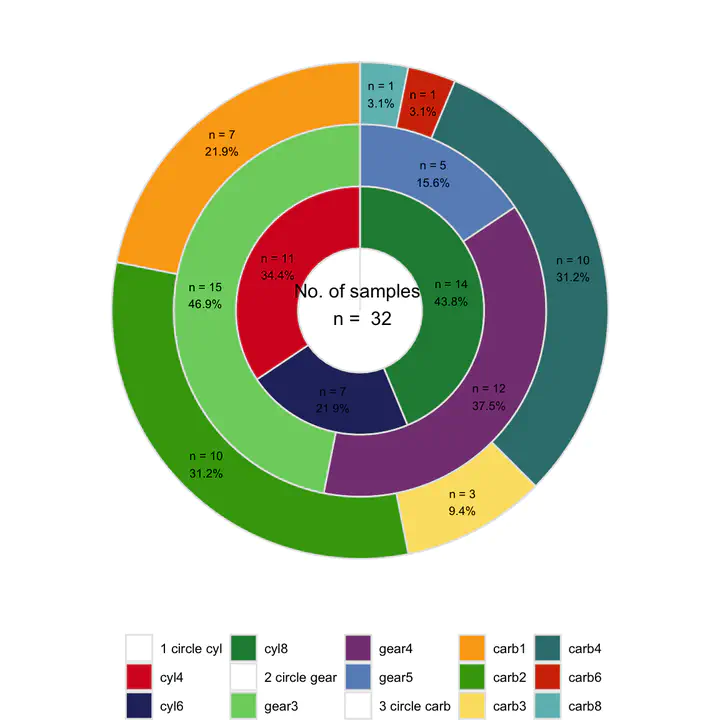
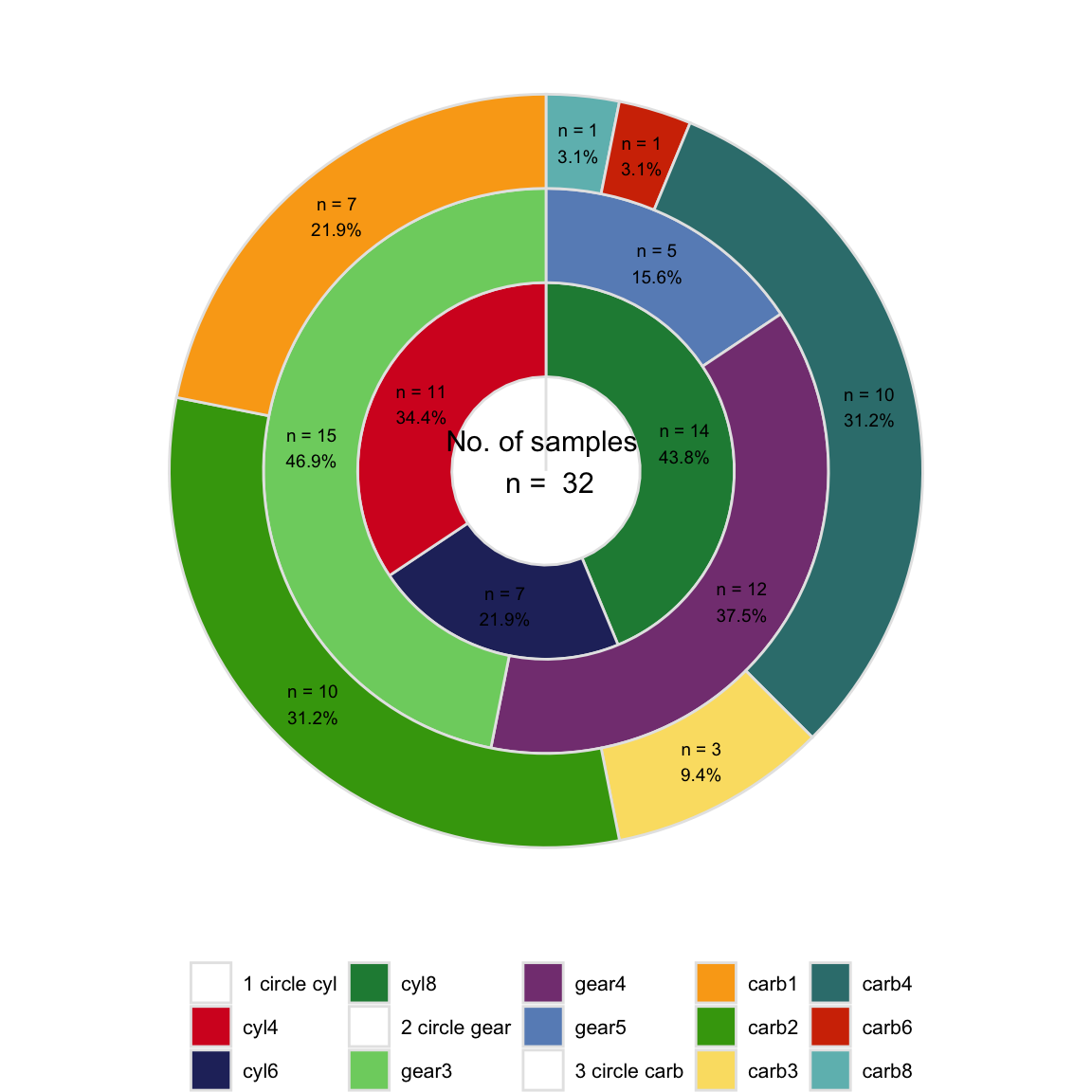
Mulitple circles chart
One circle represents one traits of dataset, should we using multiple circles to characterize the different traits?
Traits of mtcars: cyl, gear and carb
- Data preparation
data_select <- mtcars |>
dplyr::select(cyl, gear, carb) |>
mutate_if(is.numeric, as.character) |>
mutate_if(is.character, as.factor) |>
mutate(cyl = paste0("cyl", cyl),
gear = paste0("gear", gear),
carb = paste0("carb", carb))
str(data_select)
## 'data.frame': 32 obs. of 3 variables:
## $ cyl : chr "cyl6" "cyl6" "cyl4" "cyl6" ...
## $ gear: chr "gear4" "gear4" "gear4" "gear3" ...
## $ carb: chr "carb4" "carb4" "carb1" "carb1" ...
- Calculating the Proportion per group
plotdata2 <- data_select |>
dplyr::group_by(cyl, gear, carb) |>
dplyr::summarise(count = length(cyl))
head(plotdata2)
## # A tibble: 6 × 4
## # Groups: cyl, gear [5]
## cyl gear carb count
## <chr> <chr> <chr> <int>
## 1 cyl4 gear3 carb1 1
## 2 cyl4 gear4 carb1 4
## 3 cyl4 gear4 carb2 4
## 4 cyl4 gear5 carb2 2
## 5 cyl6 gear3 carb1 2
## 6 cyl6 gear4 carb4 4
Function for drawing chart
single or multiple traits for plotting
values and position of label
plotting
get_circle <- function(
inputdata,
variables = c("cyl", "gear", "carb"),
label_size = 2.5,
annotate_size = 4,
col_number = 5,
theme_legend_size = 8,
theme_strip_size = 8,
axis_size = 10,
cyl_names = c("cyl4", "cyl6","cyl8"),
cyl_cols = c("#D51F26", "#272E6A", "#208A42"),
gear_names = c("gear3", "gear4", "gear5"),
gear_cols = c("#7DD06F", "#844081", "#688EC1"),
carb_names = c("carb1", "carb2", "carb3", "carb4", "carb6", "carb8"),
carb_cols = c("#faa818", "#41a30d", "#fbdf72", "#367d7d", "#d33502", "#6ebcbc")) {
dat_list <- list()
for (i in 1:length(variables)) {
temp_var <- rlang::sym(variables[i])
temp_dat <- inputdata %>%
dplyr::select(!!temp_var, count) %>%
dplyr::group_by(!!temp_var) %>%
dplyr::summarise(count = sum(count), .groups = "drop")
dat_list[[i]] <- temp_dat
}
names(dat_list) <- variables
get_plotdata <- function(dat) {
dat_cln_list <- list()
names_new_list <- list()
colors_new_list <- list()
for (i in 1:length(dat)) {
if (names(dat)[i] == "cyl") {
cyl_cln <- dat[[i]] %>%
dplyr::select(cyl, count) %>%
dplyr::group_by(cyl) %>%
dplyr::summarise(count = sum(count)) %>%
dplyr::mutate(prop = round(count / sum(count), 3) * 100) %>%
dplyr::ungroup() %>%
dplyr::arrange(desc(cyl)) %>%
dplyr::mutate(lab.ypos = cumsum(prop) - 0.5*prop) %>%
dplyr::mutate(num_prop = paste0("n = ", count, "\n",
paste0(prop, "%")))
cyl_cln$level <- i
cyl_cln$type <- "cyl"
match_order_index1 <- sort(pmatch(unique(cyl_cln$cyl), cyl_names), decreasing = F)
cyl_names_new <- cyl_names[match_order_index1]
cyl_cols_new <- cyl_cols[match_order_index1]
cyl_cln$fill <- factor(cyl_cln$cyl, levels = cyl_names_new, labels = cyl_cols_new)
dat_cln_list[[i]] <- cyl_cln
names_new_list[[i]] <- cyl_names_new
colors_new_list[[i]] <- cyl_cols_new
} else if (names(dat)[i] == "gear") {
gear_cln <- dat[[i]] %>%
dplyr::select(gear, count) %>%
dplyr::group_by(gear) %>%
dplyr::summarise(count = sum(count)) %>%
dplyr::mutate(prop = round(count / sum(count), 3) * 100) %>%
dplyr::ungroup() %>%
dplyr::arrange(desc(gear)) %>%
dplyr::mutate(lab.ypos = cumsum(prop) - 0.5*prop) %>%
dplyr::mutate(num_prop = paste0("n = ", count, "\n",
paste0(prop, "%")))
gear_cln$level <- i
gear_cln$type <- "gear"
match_order_index2 <- sort(pmatch(unique(gear_cln$gear), gear_names), decreasing = F)
gear_names_new <- gear_names[match_order_index2]
gear_cols_new <- gear_cols[match_order_index2]
gear_cln$fill <- factor(gear_cln$gear, levels = gear_names_new, labels = gear_cols_new)
dat_cln_list[[i]] <- gear_cln
names_new_list[[i]] <- gear_names_new
colors_new_list[[i]] <- gear_cols_new
} else if (names(dat)[i] == "carb") {
carb_cln <- dat[[i]] %>%
dplyr::select(carb, count) %>%
dplyr::group_by(carb) %>%
dplyr::summarise(count = sum(count)) %>%
dplyr::mutate(prop = round(count / sum(count), 3) * 100) %>%
dplyr::ungroup() %>%
dplyr::arrange(desc(carb)) %>%
dplyr::mutate(lab.ypos = cumsum(prop) - 0.5*prop) %>%
dplyr::mutate(num_prop = paste0("n = ", count, "\n",
paste0(prop, "%")))
carb_cln$level <- i
carb_cln$type <- "carb"
match_order_index3 <- sort(pmatch(unique(carb_cln$carb), carb_names), decreasing = F)
carb_names_new <- carb_names[match_order_index3]
carb_cols_new <- carb_cols[match_order_index3]
carb_cln$fill <- factor(carb_cln$carb, levels = carb_names_new, labels = carb_cols_new)
dat_cln_list[[i]] <- carb_cln
names_new_list[[i]] <- carb_names_new
colors_new_list[[i]] <- carb_cols_new
}
}
names(dat_cln_list) <- names(dat)
names(names_new_list) <- names(dat)
names(colors_new_list) <- names(dat)
temp_name <- c()
temp_value <- c()
temp_level <- c()
temp_type <- c()
temp_num_prop <- c()
for (j in 1:length(dat_cln_list)) {
temp_name <- c(temp_name, dat_cln_list[[j]][, 1, T])
temp_value <- c(temp_value, dat_cln_list[[j]][, 2, T])
temp_level <- c(temp_level, dat_cln_list[[j]][, 6, T])
temp_type <- c(temp_type, dat_cln_list[[j]][, 7, T])
temp_num_prop <- c(temp_num_prop, dat_cln_list[[j]][, 5, T])
}
data_temp <- data.frame(name = temp_name,
value = temp_value,
level = temp_level,
type = temp_type,
num_prop = temp_num_prop)
data_parent <- data.frame(name = "Parent", value = 0, level = 0, type = NA, num_prop = NA)
dat_res <- rbind(data_parent, data_temp)
res <- list(plotdata = dat_res,
names_new = names_new_list,
colors_new = colors_new_list)
return(res)
}
# discrete variables
res_list <- get_plotdata(dat = dat_list)
plotdata <- res_list$plotdata
name_levels <- c()
name_colors <- c()
for (k in 1:length(res_list$names_new)) {
temp_name <- c(paste(k, "circle", names(res_list$names_new)[k]), res_list$names_new[[k]])
temp_color <- c("white", res_list$colors_new[[k]])
name_levels <- c(name_levels, temp_name)
name_colors <- c(name_colors, temp_color)
}
names(name_colors) <- name_levels
plotdata$name <- factor(plotdata$name, levels = name_levels)
total_num <- plotdata %>%
dplyr::pull(value) %>%
sum() / length(variables)
pl <- ggplot(plotdata, aes(x = level, y = value, fill = name)) +
geom_col(width = 1, color = "gray90", size = 0.5, position = position_stack()) +
geom_text(aes(label = num_prop), size = label_size, position = position_stack(vjust = 0.5)) +
coord_polar(theta = "y", start = 0) +
scale_x_discrete(breaks = NULL) +
scale_y_continuous(breaks = NULL) +
labs(x = NULL, y = NULL) +
scale_fill_manual(values = name_colors, drop = FALSE) +
guides(fill = guide_legend(title = NULL, ncol = col_number)) +
annotate("text", x = -0.4, y = 0,
label = paste("No. of samples", "\n n = ", total_num),
size = annotate_size) +
theme_void() +
theme(axis.title = element_text(face = "bold", size = axis_size),
axis.text = element_text(size = axis_size - 1),
text = element_text(size = axis_size - 2),
legend.position = "bottom",
legend.text = element_text(size = theme_legend_size, color = "black"),
strip.text.x = element_text(size = theme_strip_size, color = "black", face = "bold"))
return(pl)
}
- multiple circles
pl_circle <- get_circle(
inputdata = plotdata2,
variables = c("cyl", "gear", "carb"),
label_size = 2.5,
annotate_size = 4,
col_number = 5,
theme_legend_size = 8,
theme_strip_size = 8,
axis_size = 10)
pl_circle
Conclusion
Pie and donut only are fit for proportion of single variable, while multiple circles are suitable for ratio of multiple traits in dataset.
Systemic information
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.3 (2022-03-10)
## os macOS Big Sur/Monterey 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2023-07-24
## pandoc 3.1.3 @ /Users/zouhua/opt/anaconda3/bin/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## blogdown 1.18 2023-06-19 [2] CRAN (R 4.1.3)
## bookdown 0.34 2023-05-09 [2] CRAN (R 4.1.2)
## bslib 0.5.0 2023-06-09 [2] CRAN (R 4.1.3)
## cachem 1.0.8 2023-05-01 [2] CRAN (R 4.1.2)
## callr 3.7.3 2022-11-02 [2] CRAN (R 4.1.2)
## cli 3.6.1 2023-03-23 [2] CRAN (R 4.1.2)
## colorspace 2.1-0 2023-01-23 [2] CRAN (R 4.1.2)
## crayon 1.5.2 2022-09-29 [2] CRAN (R 4.1.2)
## devtools 2.4.5 2022-10-11 [2] CRAN (R 4.1.2)
## digest 0.6.33 2023-07-07 [1] CRAN (R 4.1.3)
## dplyr * 1.1.2 2023-04-20 [2] CRAN (R 4.1.2)
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.1.0)
## evaluate 0.21 2023-05-05 [2] CRAN (R 4.1.2)
## fansi 1.0.4 2023-01-22 [2] CRAN (R 4.1.2)
## farver 2.1.1 2022-07-06 [2] CRAN (R 4.1.2)
## fastmap 1.1.1 2023-02-24 [2] CRAN (R 4.1.2)
## forcats * 1.0.0 2023-01-29 [2] CRAN (R 4.1.2)
## fs 1.6.2 2023-04-25 [2] CRAN (R 4.1.2)
## generics 0.1.3 2022-07-05 [2] CRAN (R 4.1.2)
## ggplot2 * 3.4.2 2023-04-03 [2] CRAN (R 4.1.2)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.1.2)
## gtable 0.3.3 2023-03-21 [2] CRAN (R 4.1.2)
## highr 0.10 2022-12-22 [2] CRAN (R 4.1.2)
## hms 1.1.3 2023-03-21 [2] CRAN (R 4.1.2)
## htmltools 0.5.5 2023-03-23 [2] CRAN (R 4.1.2)
## htmlwidgets 1.6.2 2023-03-17 [2] CRAN (R 4.1.2)
## httpuv 1.6.11 2023-05-11 [2] CRAN (R 4.1.3)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.1.0)
## jsonlite 1.8.7 2023-06-29 [2] CRAN (R 4.1.3)
## knitr 1.43 2023-05-25 [2] CRAN (R 4.1.3)
## labeling 0.4.2 2020-10-20 [2] CRAN (R 4.1.0)
## later 1.3.1 2023-05-02 [2] CRAN (R 4.1.2)
## lifecycle 1.0.3 2022-10-07 [2] CRAN (R 4.1.2)
## lubridate * 1.9.2 2023-02-10 [2] CRAN (R 4.1.2)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.1.2)
## memoise 2.0.1 2021-11-26 [2] CRAN (R 4.1.0)
## mime 0.12 2021-09-28 [2] CRAN (R 4.1.0)
## miniUI 0.1.1.1 2018-05-18 [2] CRAN (R 4.1.0)
## munsell 0.5.0 2018-06-12 [2] CRAN (R 4.1.0)
## pillar 1.9.0 2023-03-22 [2] CRAN (R 4.1.2)
## pkgbuild 1.4.2 2023-06-26 [2] CRAN (R 4.1.3)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.1.0)
## pkgload 1.3.2.1 2023-07-08 [2] CRAN (R 4.1.3)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.1.0)
## processx 3.8.2 2023-06-30 [2] CRAN (R 4.1.3)
## profvis 0.3.8 2023-05-02 [2] CRAN (R 4.1.2)
## promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.1.0)
## ps 1.7.5 2023-04-18 [2] CRAN (R 4.1.2)
## purrr * 1.0.1 2023-01-10 [2] CRAN (R 4.1.2)
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.1.0)
## Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.1.3)
## readr * 2.1.4 2023-02-10 [2] CRAN (R 4.1.2)
## remotes 2.4.2 2021-11-30 [2] CRAN (R 4.1.0)
## rlang 1.1.1 2023-04-28 [2] CRAN (R 4.1.2)
## rmarkdown 2.23 2023-07-01 [2] CRAN (R 4.1.3)
## rstudioapi 0.15.0 2023-07-07 [2] CRAN (R 4.1.3)
## sass 0.4.6 2023-05-03 [2] CRAN (R 4.1.2)
## scales 1.2.1 2022-08-20 [2] CRAN (R 4.1.2)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.1.0)
## shiny 1.7.4.1 2023-07-06 [2] CRAN (R 4.1.3)
## stringi 1.7.12 2023-01-11 [2] CRAN (R 4.1.2)
## stringr * 1.5.0 2022-12-02 [2] CRAN (R 4.1.2)
## tibble * 3.2.1 2023-03-20 [2] CRAN (R 4.1.2)
## tidyr * 1.3.0 2023-01-24 [2] CRAN (R 4.1.2)
## tidyselect 1.2.0 2022-10-10 [2] CRAN (R 4.1.2)
## tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.1.2)
## timechange 0.2.0 2023-01-11 [2] CRAN (R 4.1.2)
## tzdb 0.4.0 2023-05-12 [2] CRAN (R 4.1.3)
## urlchecker 1.0.1 2021-11-30 [2] CRAN (R 4.1.0)
## usethis 2.2.2 2023-07-06 [2] CRAN (R 4.1.3)
## utf8 1.2.3 2023-01-31 [2] CRAN (R 4.1.2)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.1.3)
## withr 2.5.0 2022-03-03 [2] CRAN (R 4.1.2)
## xfun 0.39 2023-04-20 [2] CRAN (R 4.1.2)
## xtable 1.8-4 2019-04-21 [2] CRAN (R 4.1.0)
## yaml 2.3.7 2023-01-23 [2] CRAN (R 4.1.2)
##
## [1] /Users/zouhua/Library/R/x86_64/4.1/library
## [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────